TCGA and ESTIMATE data mining to identify potential prognostic biomarkers in HCC patients
- PMID: 33177245
- PMCID: PMC7695391
- DOI: 10.18632/aging.103943
TCGA and ESTIMATE data mining to identify potential prognostic biomarkers in HCC patients
Abstract
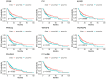
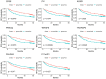
Hepatocellular carcinoma (HCC) is an aggressive form of cancer characterized by a high recurrence rate following resection. Studies have implicated stromal and immune cells, which form part of the tumor microenvironment, as significant contributors to the poor prognoses of HCC patients. In the present study, we first downloaded gene expression datasets for HCC patients from The Cancer Genome Atlas database and categorized the patients into low and high stromal or immune score groups. By comparing those groups, we identified differentially expressed genes significantly associated with HCC prognosis. The Gene Ontology database was then used to perform functional enrichment analysis, and the STRING network database was used to construct protein-protein interaction networks. Our results show that most of the differentially expressed genes were involved in immune processes and responses and the plasma membrane. Those results were then validated using another a dataset from a HCC cohort in the Gene Expression Omnibus database and in 10 pairs of HCC tumor tissue and adjacent nontumor tissue. These findings enabled us to identify several tumor microenvironment-related genes that associate with HCC prognosis, and some those appear to have the potential to serve as HCC biomarkers.
Keywords: GEO; TCGA; disease-free survival; immune scores; tumor microenvironment.
Conflict of interest statement
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

