A comparative genomics multitool for scientific discovery and conservation
- PMID: 33177664
- PMCID: PMC7759459
- DOI: 10.1038/s41586-020-2876-6
A comparative genomics multitool for scientific discovery and conservation
Abstract
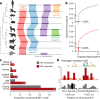
The Zoonomia Project is investigating the genomics of shared and specialized traits in eutherian mammals. Here we provide genome assemblies for 131 species, of which all but 9 are previously uncharacterized, and describe a whole-genome alignment of 240 species of considerable phylogenetic diversity, comprising representatives from more than 80% of mammalian families. We find that regions of reduced genetic diversity are more abundant in species at a high risk of extinction, discern signals of evolutionary selection at high resolution and provide insights from individual reference genomes. By prioritizing phylogenetic diversity and making data available quickly and without restriction, the Zoonomia Project aims to support biological discovery, medical research and the conservation of biodiversity.
Conflict of interest statement
L.G. is a co-founder of, equity owner in and chief technical officer at Fauna Bio Incorporated.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- 1UH2AG064706/NH/NIH HHS/United States
- 5R01HG010485/NH/NIH HHS/United States
- P01 AG047200/AG/NIA NIH HHS/United States
- R01 HG008742/HG/NHGRI NIH HHS/United States
- 200517/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- U24 HG010136/HG/NHGRI NIH HHS/United States
- R01 HG010485/HG/NHGRI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- 5U24HG010136/NH/NIH HHS/United States
- 5P01AG047200/NH/NIH HHS/United States
- 2U41HG007234/NH/NIH HHS/United States
- U41 HG007234/HG/NHGRI NIH HHS/United States
- R01 HG002939/HG/NHGRI NIH HHS/United States
- UH2 AG064706/AG/NIA NIH HHS/United States

