Flow-Induced Transcriptomic Remodeling of Endothelial Cells Derived From Human Induced Pluripotent Stem Cells
- PMID: 33178051
- PMCID: PMC7593792
- DOI: 10.3389/fphys.2020.591450
Flow-Induced Transcriptomic Remodeling of Endothelial Cells Derived From Human Induced Pluripotent Stem Cells
Abstract
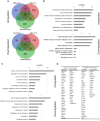
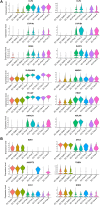
The vascular system is essential for the development and function of all organs and tissues in our body. The molecular signature and phenotype of endothelial cells (EC) are greatly affected by blood flow-induced shear stress, which is a vital component of vascular development and homeostasis. Recent advances in differentiation of ECs from human induced pluripotent stem cells (hiPSC) have enabled development of in vitro experimental models of the vasculature containing cells from healthy individuals or from patients harboring genetic variants or diseases of interest. Here we have used hiPSC-derived ECs and bulk- and single-cell RNA sequencing to study the effect of flow on the transcriptomic landscape of hiPSC-ECs and their heterogeneity. We demonstrate that hiPS-ECs are plastic and they adapt to flow by expressing known flow-induced genes. Single-cell RNA sequencing showed that flow induced a more homogenous and homeostatically more stable EC population compared to static cultures, as genes related to cell polarization, barrier formation and glucose and fatty acid transport were induced. The hiPS-ECs increased both arterial and venous markers when exposed to flow. Interestingly, while in general there was a greater increase in the venous markers, one cluster with more arterial-like hiPS-ECs was detected. Single-cell RNA sequencing revealed that not all hiPS-ECs are similar even after sorting, but exposing them to flow increases their homogeneity. Since hiPS-ECs resemble immature ECs and demonstrate high plasticity in response to flow, they provide an excellent model to study vascular development.
Keywords: RNA sequencing; endothelial cells; flow; induced pluripotent stem cells; shear stress; single-cell RNA sequencing.
Copyright © 2020 Helle, Ampuja, Antola and Kivelä.
Figures





Similar articles
-
Functional Characterization of Human Induced Pluripotent Stem Cell-Derived Endothelial Cells.Int J Mol Sci. 2022 Jul 31;23(15):8507. doi: 10.3390/ijms23158507. Int J Mol Sci. 2022. PMID: 35955642 Free PMC article.
-
Human induced pluripotent stem cell-derived endothelial cells exhibit functional heterogeneity.Am J Transl Res. 2013;5(1):21-35. Epub 2013 Jan 21. Am J Transl Res. 2013. PMID: 23390563 Free PMC article.
-
Functionality of endothelial cells and pericytes from human pluripotent stem cells demonstrated in cultured vascular plexus and zebrafish xenografts.Arterioscler Thromb Vasc Biol. 2014 Jan;34(1):177-86. doi: 10.1161/ATVBAHA.113.302598. Epub 2013 Oct 24. Arterioscler Thromb Vasc Biol. 2014. PMID: 24158517
-
Cellular heterogeneity and stem cells of vascular endothelial cells in blood vessel formation and homeostasis: Insights from single-cell RNA sequencing.Front Cell Dev Biol. 2023 Mar 21;11:1146399. doi: 10.3389/fcell.2023.1146399. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37025170 Free PMC article. Review.
-
Environmental Specification of Pluripotent Stem Cell Derived Endothelial Cells Toward Arterial and Venous Subtypes.Front Bioeng Biotechnol. 2019 Jun 14;7:143. doi: 10.3389/fbioe.2019.00143. eCollection 2019. Front Bioeng Biotechnol. 2019. PMID: 31259171 Free PMC article. Review.
Cited by
-
New Technologies With Increased Precision Improve Understanding of Endothelial Cell Heterogeneity in Cardiovascular Health and Disease.Front Cell Dev Biol. 2021 Aug 27;9:679995. doi: 10.3389/fcell.2021.679995. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34513826 Free PMC article. Review.
-
Organotypic stromal cells impact endothelial cell transcriptome in 3D microvessel networks.Sci Rep. 2022 Nov 28;12(1):20434. doi: 10.1038/s41598-022-24013-y. Sci Rep. 2022. PMID: 36443378 Free PMC article.
-
Remodeling of self-assembled microvascular networks under long term flow.bioRxiv [Preprint]. 2025 Mar 18:2025.03.17.643791. doi: 10.1101/2025.03.17.643791. bioRxiv. 2025. PMID: 40166169 Free PMC article. Preprint.
-
Translational potential of hiPSCs in predictive modeling of heart development and disease.Birth Defects Res. 2022 Oct 1;114(16):926-947. doi: 10.1002/bdr2.1999. Epub 2022 Mar 9. Birth Defects Res. 2022. PMID: 35261209 Free PMC article. Review.
-
Patterned Arteriole-Scale Vessels Enhance Engraftment, Perfusion, and Vessel Branching Hierarchy of Engineered Human Myocardium for Heart Regeneration.Cells. 2023 Jun 23;12(13):1698. doi: 10.3390/cells12131698. Cells. 2023. PMID: 37443731 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

