Cryptosporidium parvum Infection Depletes Butyrate Producer Bacteria in Goat Kid Microbiome
- PMID: 33178145
- PMCID: PMC7596689
- DOI: 10.3389/fmicb.2020.548737
Cryptosporidium parvum Infection Depletes Butyrate Producer Bacteria in Goat Kid Microbiome
Abstract
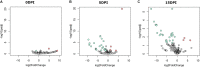
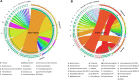
Cryptosporidium parvum is an important apicomplexan parasite infecting ruminants and humans. We characterized the impact of C. parvum infection on the goat kid microbiome. C. parvum was orally administered to parasite-naïve goats, and infection was monitored for 26 days in fecal samples using immunofluorescence assay and qPCR tests. Age-matched goats served as uninfected controls. A reduction in body weight gain, diarrhea, and dehydration were observed in infected goats compared to the uninfected controls. Infection decreased the bacterial diversity 5 days post-infection (dpi), but this parameter recovered at 15 dpi. The infection altered the relative abundance of several taxa. A total of 38 taxa displayed significant differences in abundance between control and infected goats at both 5 and 15 dpi. Co-occurrence network analysis revealed that the infection resulted in a differential pattern of taxa interactions and that C. parvum infection increased the relative abundance of specific taxa. The 16S data set was used for metagenome predictions using the software package PICRUSt2. As many as 34 and 40 MetaCyc pathways (from 387 total) were significantly affected by the infection at 5 and 15 dpi, respectively. Notably, C. parvum decreased the abundance of butyrate-producing pathways in bacteria. Low levels of butyrate may increase mucosal inflammation and tissue repair. Our results suggest that the gut inflammation induced by C. parvum infection is associated with the reduction of butyrate-producing bacteria. This insight could be the basis for the development of novel control strategies to improve animal health.
Keywords: 16S; diarrhea; dysbiosis; functional traits; gut microbiome; resilience 3.
Copyright © 2020 Mammeri, Obregón, Chevillot, Polack, Julien, Pollet, Cabezas-Cruz and Adjou.
Figures





References
-
- Bastian M., Jacomy M. (2009). “Gephi: an open source software for exploring and manipulating networks gephi: an open source software for exploring and manipulating networks,” in Proceedings of the Third International Conference on Weblogs and Social Media, ICWSM 2009, (San Jose, CA: ), 4–6. 10.13140/2.1.1341.1520 - DOI
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57 289–300. 10.1111/j.2517-6161.1995.tb02031.x - DOI
LinkOut - more resources
Full Text Sources
Research Materials

