Occurrence of Yeasts in White-Brined Cheeses: Methodologies for Identification, Spoilage Potential and Good Manufacturing Practices
- PMID: 33178163
- PMCID: PMC7593773
- DOI: 10.3389/fmicb.2020.582778
Occurrence of Yeasts in White-Brined Cheeses: Methodologies for Identification, Spoilage Potential and Good Manufacturing Practices
Abstract
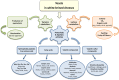
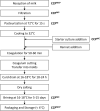
Yeasts are generally recognized as contaminants in the production of white-brined cheeses, such as Feta and Feta-type cheeses. The most predominant yeasts species are Debaryomyces hansenii, Geotrichum candidum, Kluyveromyces marxianus, Kluyveromyces lactis, Rhodotorula mucilaginosa, and Trichosporon spp. Although their spoilage potential varies at both species and strain levels, yeasts will, in case of excessive growth, present a microbiological hazard, effecting cheese quality. To evaluate the hazard and trace routes of contamination, the exact taxonomic classification of yeasts is required. Today, identification of dairy yeasts is mainly based on DNA sequencing, various genotyping techniques, and, to some extent, advanced phenotypic identification technologies. Even though these technologies are state of the art at the scientific level, they are only hardly implemented at the industrial level. Quality defects, caused by yeasts in white-brined cheese, are mainly linked to enzymatic activities and metabolism of fermentable carbohydrates, leading to production of metabolites (CO2, fatty acids, volatile compounds, amino acids, sulfur compounds, etc.) and resulting in off-flavors, texture softening, discoloration, and swelling of cheese packages. The proliferation of spoilage yeast depends on maturation and storage conditions at each specific dairy, product characteristics, nutrients availability, and interactions with the co-existing microorganisms. To prevent and control yeast contamination, different strategies based on the principles of HACCP and Good Manufacturing Practice (GMP) have been introduced in white-brined cheese production. These strategies include milk pasteurization, refrigeration, hygienic sanitation, air filtration, as well as aseptic and modified atmosphere packaging. Though a lot of research has been dedicated to yeasts in dairy products, the role of yeast contaminants, specifically in white-brined cheeses, is still insufficiently understood. This review aims to summarize the current knowledge on the identification of contaminant yeasts in white-brined cheeses, their occurrence and spoilage potential related to different varieties of white-brined cheeses, their interactions with other microorganisms, as well as guidelines used by dairies to prevent cheese contamination.
Keywords: GMP; microbial interactions; off-flavors; spoilage yeasts; white-brined cheese; yeast identification.
Copyright © 2020 Geronikou, Srimahaeak, Rantsiou, Triantafillidis, Larsen and Jespersen.
Figures

Similar articles
-
Occurrence and Identification of Yeasts in Production of White-Brined Cheese.Microorganisms. 2022 May 24;10(6):1079. doi: 10.3390/microorganisms10061079. Microorganisms. 2022. PMID: 35744597 Free PMC article.
-
Diversity and succession of contaminating yeasts in white-brined cheese during cold storage.Food Microbiol. 2023 Aug;113:104266. doi: 10.1016/j.fm.2023.104266. Epub 2023 Mar 20. Food Microbiol. 2023. PMID: 37098422
-
Yeasts in different types of cheese.AIMS Microbiol. 2021 Nov 8;7(4):447-470. doi: 10.3934/microbiol.2021027. eCollection 2021. AIMS Microbiol. 2021. PMID: 35071942 Free PMC article. Review.
-
Growth Capacity of a Novel Potential Probiotic Lactobacillus paracasei K5 Strain Incorporated in Industrial White Brined Cheese as an Adjunct Culture.J Food Sci. 2018 Mar;83(3):723-731. doi: 10.1111/1750-3841.14079. Epub 2018 Feb 23. J Food Sci. 2018. PMID: 29473955
-
Cheese yeasts.Yeast. 2019 Mar;36(3):129-141. doi: 10.1002/yea.3368. Epub 2019 Jan 24. Yeast. 2019. PMID: 30512214 Review.
Cited by
-
Occurrence and Identification of Yeasts in Production of White-Brined Cheese.Microorganisms. 2022 May 24;10(6):1079. doi: 10.3390/microorganisms10061079. Microorganisms. 2022. PMID: 35744597 Free PMC article.
-
Study of the Microbiome of the Cretan Sour Cream Staka Using Amplicon Sequencing and Shotgun Metagenomics and Isolation of Novel Strains with an Important Antimicrobial Potential.Foods. 2024 Apr 8;13(7):1129. doi: 10.3390/foods13071129. Foods. 2024. PMID: 38611432 Free PMC article.
-
Microbiological Assessment of Dairy Products Produced by Small-Scale Dairy Producers in Serbia.Foods. 2024 May 8;13(10):1456. doi: 10.3390/foods13101456. Foods. 2024. PMID: 38790756 Free PMC article.
-
Linking microbial contamination to food spoilage and food waste: the role of smart packaging, spoilage risk assessments, and date labeling.Front Microbiol. 2023 Jun 22;14:1198124. doi: 10.3389/fmicb.2023.1198124. eCollection 2023. Front Microbiol. 2023. PMID: 37426008 Free PMC article. Review.
-
Omics Approaches to Assess Flavor Development in Cheese.Foods. 2022 Jan 11;11(2):188. doi: 10.3390/foods11020188. Foods. 2022. PMID: 35053920 Free PMC article. Review.
References
-
- Aday S., Karagul Yuceer Y. (2014). Physicochemical and sensory properties of Mihalic cheese. Int. J. Food Prop. 17 2207–2227. 10.1080/10942912.2013.790904 - DOI
-
- Al-Dabbas M. M., Saleh M., Abu-Ghoush M. H., Al-Ismail K., Osaili T. (2014). Influence of storage, brine concentration and in-container heat treatment on the stability of white brined Nabulsi cheese. Int. J. Dairy Technol. 67 427–436. 10.1111/1471-0307.12139 - DOI
-
- Al-Gamal M. S., Ibrahim G. A., Sharaf O. M., Radwan A. A., Dabiza N. M., Youssef A. M., et al. (2019). The protective potential of selected lactic acid bacteria against the most common contaminants in various types of cheese in Egypt. Heliyon 5:e01362. 10.1016/j.heliyon.2019.e01362 - DOI - PMC - PubMed
-
- Aliakbarlu J., Alizadeh M., Razavi-Rohani S. M., Agh N. (2011). Biogenic amines in Iranian white brine cheese: modelling and optimisation of processing factors. Int. J. Dairy Technol. 64 417–424. 10.1111/j.1471-0307.2011.00681.x - DOI
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

