Functional Cross-Talk of MbtH-Like Proteins During Thaxtomin Biosynthesis in the Potato Common Scab Pathogen Streptomyces scabiei
- PMID: 33178168
- PMCID: PMC7593251
- DOI: 10.3389/fmicb.2020.585456
Functional Cross-Talk of MbtH-Like Proteins During Thaxtomin Biosynthesis in the Potato Common Scab Pathogen Streptomyces scabiei
Abstract
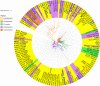
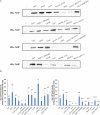
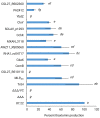
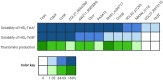
Thaxtomin A is a potent phytotoxin that serves as the principle pathogenicity determinant of the common scab pathogen, Streptomyces scabiei, and is also a promising natural herbicide for agricultural applications. The biosynthesis of thaxtomin A involves the non-ribosomal peptide synthetases (NRPSs) TxtA and TxtB, and an MbtH-like protein (MLP), TxtH, which may function as a chaperone by promoting the proper folding of the two NRPS enzymes in S. scabiei. MLPs are required for the proper function of many NRPS enzymes in bacteria, and they are often capable of interacting with NRPSs from different biosynthetic pathways, though the mechanism by which this occurs is still poorly understood. To gain additional insights into MLP functional cross-talk, we conducted a broad survey of MLPs from diverse phylogenetic lineages to determine if they could functionally replace TxtH. The MLPs were assessed using a protein solubility assay to determine whether they could promote the soluble expression of the TxtA and TxtB adenylation domains. In addition, the MLPs were tested for their ability to restore thaxtomin production in a S. scabiei mutant that lacked TxtH and other endogenous MLPs. Our results showed that the MLPs investigated vary in their ability to exhibit functional cross-talk with TxtH, with two of the MLPs being unable to compensate for the loss of TxtH in the assays performed. The ability of an MLP to serve as a functional partner for the thaxtomin NRPS was not correlated with its overall amino acid similarity with TxtH, but instead with the presence of highly conserved residues. In silico structural analysis of TxtH in association with the TxtA and TxtB adenylation domains revealed that several such residues are situated at the predicted interaction interface, suggesting that they might be critical for promoting functional interactions between MLPs and the thaxtomin NRPS enzymes. Overall, our study provides additional insights into the mechanism of MLP cross-talk, and it enhances our understanding of the thaxtomin biosynthetic machinery. It is anticipated that our findings will have useful applications for both the control of common scab disease and the commercial production of thaxtomin A for agricultural use.
Keywords: Streptomyces; non-ribosomal peptides; phytotoxin; plant pathogen; specialized metabolism; thaxtomin.
Copyright © 2020 Li, Tahlan and Bignell.
Figures


Similar articles
-
TxtH is a key component of the thaxtomin biosynthetic machinery in the potato common scab pathogen Streptomyces scabies.Mol Plant Pathol. 2019 Oct;20(10):1379-1393. doi: 10.1111/mpp.12843. Epub 2019 Jul 7. Mol Plant Pathol. 2019. PMID: 31282068 Free PMC article.
-
An Orphan MbtH-Like Protein Interacts with Multiple Nonribosomal Peptide Synthetases in Myxococcus xanthus DK1622.J Bacteriol. 2018 Oct 10;200(21):e00346-18. doi: 10.1128/JB.00346-18. Print 2018 Nov 1. J Bacteriol. 2018. PMID: 30126939 Free PMC article.
-
The cellobiose sensor CebR is the gatekeeper of Streptomyces scabies pathogenicity.mBio. 2015 Feb 24;6(2):e02018. doi: 10.1128/mBio.02018-14. mBio. 2015. PMID: 25714708 Free PMC article.
-
Evolution of plant pathogenicity in Streptomyces.Annu Rev Phytopathol. 2006;44:469-87. doi: 10.1146/annurev.phyto.44.032905.091147. Annu Rev Phytopathol. 2006. PMID: 16719719 Review.
-
Chemical Strategies for Visualizing and Analyzing Endogenous Nonribosomal Peptide Synthetase (NRPS) Megasynthetases.Chembiochem. 2019 Aug 16;20(16):2032-2040. doi: 10.1002/cbic.201900186. Epub 2019 Jul 22. Chembiochem. 2019. PMID: 31134733 Review.
Cited by
-
Identification of the Biosynthetic Gene Cluster of New Piperazic Acid-Containing Lipopeptides with Cytotoxic Activity in the Genome of Marine Streptomyces PHM034.Metabolites. 2023 Oct 18;13(10):1091. doi: 10.3390/metabo13101091. Metabolites. 2023. PMID: 37887416 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

