Molecular Pap Smear: Validation of HPV Genotype and Host Methylation Profiles of ADCY8, CDH8, and ZNF582 as a Predictor of Cervical Cytopathology
- PMID: 33178175
- PMCID: PMC7593258
- DOI: 10.3389/fmicb.2020.595902
Molecular Pap Smear: Validation of HPV Genotype and Host Methylation Profiles of ADCY8, CDH8, and ZNF582 as a Predictor of Cervical Cytopathology
Abstract
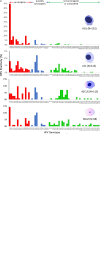
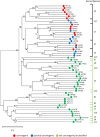
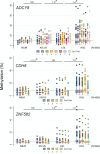
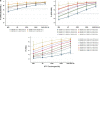
Primary high-risk Human Papillomavirus (hrHPV) screening has recently become an accepted standalone or co-test with conventional cytology. Unfortunately, hrHPV singularly lacks specificity for cytopathological grade. However, mechanisms and markers of evolving virus-host interactions at the epigenome level may be harnessed as a better predictor of carcinogenesis. This study aimed to validate and expand the clinical performance of a multiparametric biomarker panel, referred to as the "Molecular Pap smear" based, on HPV genotype and ADCY8, CDH8 and ZNF582 CpG-methylation as a predictive classifier of cervical cytology. This prospective, cross-sectional study used an independent cohort of residual liquid-based cytology for HPV genotyping and epigenetic analysis. Extracted DNA underwent parallel PCR using 3 primer sets for HPV DNA amplification. HPV-infected samples were genotyped by Sanger sequencing. Promoter methylation levels of 3 tumor suppressor genes were quantified by bisulfite-pyrosequencing of genomic DNA on the newest high-resolution PyroMark Q48 platform. Logistic model performance was compared, and model parameters were used to predict and classify binary cytological outcomes. A total of 883 samples were analyzed. HPV DNA positivity correlated with worsening grade: 125/237 (53%) NILM; 136/235 (58%) ASCUS; 222/229 (97%) LSIL; and 157/182 (86%) HSIL samples. The proportion of carcinogenic HPV-types in PCR-positive sequenceable samples correlated with worsening grade: NILM 34/98 (35%); ASCUS 50/113 (44%); LSIL 92/214 (43%); HSIL 129/152 (85%). Additionally, ADCY8, CDH8, and ZNF582 methylation levels increased in direct correlation with worsening grade. Overall, the multi-marker modeling parameters predicted binarized cytological outcomes better than HPV-type alone with significantly higher area under the receiver operator curve (AUC)s, respectively: NILM vs. > NILM (AUC 0.728 vs. 0.709); NILM/ASCUS vs. LSIL/HSIL (AUC 0.805 vs. 0.776); and <HSIL vs. HSIL (AUC 0.830 vs. 0.761). Our expanded findings validated the multivariable prediction model developed for cytological classification. The sequencing-based "Molecular Pap smear" outperformed HPV-type alone in predicting four grades of cervical cytology. Additional host epigenetic markers that evolved with disease progression decidedly contributed to the overall classification accuracy.
Keywords: DNA methylation; carcinogenesis; epigenetic modification; evolution; host-pathogen interactions; human papillomavirus infection; pap smear; pyrosequencing.
Copyright © 2020 Shen-Gunther, Xia, Stacey and Asusta.
Figures


References
-
- Bosch F. X., Broker T. R., Forman D., Moscicki A. B., Gillison M. L., Doorbar J. (2013). ‘Comprehensive Control of HPV Infections and Related Diseases’ Vaccine Volume 30, Supplement 5, 2012. Comprehensive control of human papillomavirus infections and related diseases. Vaccine 31(Suppl. 8), I1–I31. 10.1016/j.vaccine.2013.07.026 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

