T-Cell Hyperactivation and Paralysis in Severe COVID-19 Infection Revealed by Single-Cell Analysis
- PMID: 33178221
- PMCID: PMC7596772
- DOI: 10.3389/fimmu.2020.589380
T-Cell Hyperactivation and Paralysis in Severe COVID-19 Infection Revealed by Single-Cell Analysis
Abstract
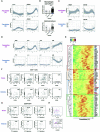
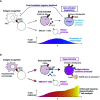
Severe COVID-19 patients show various immunological abnormalities including T-cell reduction and cytokine release syndrome, which can be fatal and is a major concern of the pandemic. However, it is poorly understood how T-cell dysregulation can contribute to the pathogenesis of severe COVID-19. Here we show single cell-level mechanisms for T-cell dysregulation in severe COVID-19, demonstrating new pathogenetic mechanisms of T-cell activation and differentiation underlying severe COVID-19. By in silico sorting CD4+ T-cells from a single cell RNA-seq dataset, we found that CD4+ T-cells were highly activated and showed unique differentiation pathways in the lung of severe COVID-19 patients. Notably, those T-cells in severe COVID-19 patients highly expressed immunoregulatory receptors and CD25, whilst repressing the expression of FOXP3. Furthermore, we show that CD25+ hyperactivated T-cells differentiate into multiple helper T-cell lineages, showing multifaceted effector T-cells with Th1 and Th2 characteristics. Lastly, we show that CD25-expressing hyperactivated T-cells produce the protease Furin, which facilitates the viral entry of SARS-CoV-2. Collectively, CD4+ T-cells from severe COVID-19 patients are hyperactivated and FOXP3-mediated negative feedback mechanisms are impaired in the lung, which may promote immunopathology. Therefore, our study proposes a new model of T-cell hyperactivation and paralysis that drives immunopathology in severe COVID-19.
Keywords: CD25; COVID-19; FOXP3; Furin; SARS-CoV-2; T-cells; regulatory T-cells (Tregs); single cell RNA-seq.
Copyright © 2020 Kalfaoglu, Almeida-Santos, Tye, Satou and Ono.
Figures



References
-
- Glowacka I, Bertram S, Muller M, Allen P, Soilleux E, Pfefferle S, et al. Evidence that TMPRSS2 Activates the Severe Acute Respiratory Syndrome Coronavirus Spike Protein for Membrane Fusion and Reduces Viral Control by the Humoral Immune Response. J Virol (2011) 85:4122–34. 10.1128/jvi.02232-10 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

