Molecular Phylogenetic Analysis of 16S rRNA Sequences Identified Two Lineages of Helicobacter pylori Strains Detected from Different Regions in Sudan Suggestive of Differential Evolution
- PMID: 33178282
- PMCID: PMC7609147
- DOI: 10.1155/2020/8825718
Molecular Phylogenetic Analysis of 16S rRNA Sequences Identified Two Lineages of Helicobacter pylori Strains Detected from Different Regions in Sudan Suggestive of Differential Evolution
Abstract
Background: Helicobacter pylori (H. pylori) is ubiquitous among humans and one of the best-studied examples of an intimate association between bacteria and humans. Phylogeny and Phylogeography of H. pylori strains are known to mirror human migration patterns and reflect significant demographic events in human prehistory. In this study, we analyzed the molecular evolution of H. pylori strains detected from different tribes and regions of Sudan using 16S rRNA gene and the phylogenetic approach. Materials and methods. A total of 75 gastric biopsies were taken from patients who had been referred for endoscopy from different regions of Sudan. The DNA extraction was performed by using the guanidine chloride method. Two sets of primers (universal and specific for H. pylori) were used to amplify the 16S ribosomal gene. Sanger sequencing was applied, and the resulted sequences were matched with the sequences of the National Center for Biotechnology Information (NCBI) nucleotide database. The evolutionary aspects were analyzed using MEGA7 software.
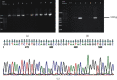
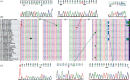
Results: Molecular detection of H. pylori has shown that 28 (37.33%) of the patients were positive for H. pylori and no significant differences were found in sociodemographic characteristics, endoscopy series, and H. pylori infection. Nucleotide variations were observed at five nucleotide positions (positions 219, 305, 578, 741, and 763-764), and one insertion mutation (750_InsC_751) was present in sixty-seven percent (7/12) of our strains. These six mutations were detected in regions of the 16S rRNA not closely associated with either tetracycline or tRNA binding sites; 66.67% of them were located in the central domain of 16S rRNA. The phylogenetic analysis of 16S rRNA sequences identified two lineages of H. pylori strains detected from different regions in Sudan. The presence of Sudanese H. pylori strains resembling Hungarian H. pylori strains could reflect the migration of Hungarian people to Sudan or vice versa.
Conclusion: This finding emphasizes the significance of studying the phylogeny of H. pylori strains as a discriminatory tool to mirror human migration patterns. In addition, the 16S rRNA gene amplification method was found useful for bacterial identification and phylogeny.
Copyright © 2020 Abeer Babiker Idris et al.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures

References
-
- Munoz-Ramirez Z. Y., Mendez-Tenorio A., Kato I., et al. Whole genome sequence and phylogenetic analysis show Helicobacter pylori strains from Latin America have followed a unique evolution pathway. Frontiers in Cellular and Infection Microbiology. 2017;7 doi: 10.3389/fcimb.2017.00050. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

