Development and Validation of Autophagy-Related Gene Signature and Nomogram for Predicting Survival in Oral Squamous Cell Carcinoma
- PMID: 33178587
- PMCID: PMC7596585
- DOI: 10.3389/fonc.2020.558596
Development and Validation of Autophagy-Related Gene Signature and Nomogram for Predicting Survival in Oral Squamous Cell Carcinoma
Abstract
Background: Autophagy, a highly conserved self-digesting process, has been deeply involved in the development and progression of oral squamous cell carcinoma (OSCC). However, the prognostic value of autophagy-related genes (ARGs) for OSCC still remains unclear. Our study set out to develop a multigene expression signature based on ARGs for individualized prognosis assessment in OSCC patients.
Methods: Based on The Cancer Genome Atlas (TCGA) database, we identified prognosis-related ARGs through univariate COX regression analysis. Then we performed the least absolute shrinkage and selection operator (LASSO) regression analysis to identify an optimal autophagy-related multigene signature with the subsequent validation in testing set, GSE41613 and GSE42743 datasets.
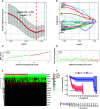
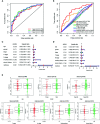
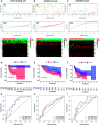
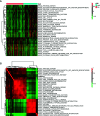
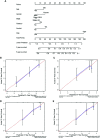
Results: We identified 36 prognosis-related ARGs for OSCC. Subsequently, the multigene signature based on 13 prognostic ARGs was constructed and successfully divided OSCC patients into low and high-risk groups with significantly different overall survival in TCGA training set (p < 0.0001). The autophagy signature remained as an independent prognostic factor for OSCC in univariate and multivariate Cox regression analyses. The area under the curve (AUC) values of the receiver operating characteristic (ROC) curves for 1, 3, and 5-year survival were 0.758, 0.810, 0.798, respectively. Then the gene signature was validated in TCGA testing set, GSE41613 and GSE42743 datasets. Moreover, Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis, and single-sample gene set enrichment analysis (ssGSEA) revealed the underlying biological characteristics and signaling pathways associated with this signature in OSCC. Finally, we constructed a nomogram by combining the gene signature with multiple clinical parameters (age, gender, TNM-stage, tobacco, and alcohol history). The concordance index (C-index) and calibration plots demonstrated favorable predictive performance of our nomogram.
Conclusion: In summary, we identified and verified a 13-ARGs prognostic signature and nomogram, which provide individualized prognosis evaluation and show insight for potential therapeutic targets for OSCC.
Keywords: The Cancer Genome Atlas; autophagy-related genes; gene signature; nomogram; oral squamous cell carcinoma; prognosis.
Copyright © 2020 Hou, Cai, Zhu, Huang, Song and Hou.
Figures



References
LinkOut - more resources
Full Text Sources

