DNA Methylation-Based Panel Predicts Survival of Patients With Clear Cell Renal Cell Carcinoma and Its Correlations With Genomic Metrics and Tumor Immune Cell Infiltration
- PMID: 33178689
- PMCID: PMC7593608
- DOI: 10.3389/fcell.2020.572628
DNA Methylation-Based Panel Predicts Survival of Patients With Clear Cell Renal Cell Carcinoma and Its Correlations With Genomic Metrics and Tumor Immune Cell Infiltration
Abstract
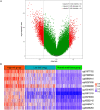
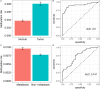
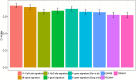
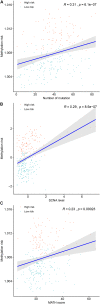
DNA methylation based prognostic factor for patients with clear cell renal cell carcinoma (ccRCC) remains unclear. In the present study, we identified survival-related DNA methylation sites based on the differentially methylated DNA CpG sites between normal renal tissue and ccRCC. Then, these survival-related DNA methylation sites were included into an elastic net regularized Cox proportional hazards regression (CoxPH) model to build a DNA methylation-based panel, which could stratify patients into different survival groups with excellent accuracies in the training set and test set. External validation suggested that the DNA methylation-based panel could effectively distinguish normal controls from tumor samples and classify patients into metastasis group and non-metastasis group. The nomogram containing DNA methylation-based panel was reliable in clinical settings. Higher total mutation number, SCNA level, and MATH score were associated with higher methylation risk. The innate immune, ratio between CD8+T cell versus Treg cell as well as Th17 cell versus Th2 cell were significantly decreased in high methylation risk group. In inclusion, we developed a DNA methylation-based panel which might be independent prognostic factor in ccRCC. Patients with higher methylation risk were associated genomic alteration and poor immune microenvironment.
Keywords: clear cell renal cell carcinoma; elastic net; methylation CpG island; prognostication panel; survival analysis.
Copyright © 2020 Liu, Ju, Chen, Liu, Li and Wang.
Figures


Similar articles
-
A DNA Methylation-Based Panel for the Prognosis and Dagnosis of Patients With Breast Cancer and Its Mechanisms.Front Mol Biosci. 2020 Jul 7;7:118. doi: 10.3389/fmolb.2020.00118. eCollection 2020. Front Mol Biosci. 2020. PMID: 32733914 Free PMC article.
-
DNA methylation status defines clinicopathological parameters including survival for patients with clear cell renal cell carcinoma (ccRCC).Tumour Biol. 2016 Aug;37(8):10219-28. doi: 10.1007/s13277-016-4893-5. Epub 2016 Jan 30. Tumour Biol. 2016. PMID: 26831665
-
Identification and Comprehensive Validation of a DNA Methylation-Driven Gene-Based Prognostic Model for Clear Cell Renal Cell Carcinoma.DNA Cell Biol. 2020 Oct;39(10):1799-1812. doi: 10.1089/dna.2020.5601. Epub 2020 Jul 21. DNA Cell Biol. 2020. PMID: 32716214
-
LAT, HOXD3 and NFE2L3 identified as novel DNA methylation-driven genes and prognostic markers in human clear cell renal cell carcinoma by integrative bioinformatics approaches.J Cancer. 2019 Oct 22;10(26):6726-6737. doi: 10.7150/jca.35641. eCollection 2019. J Cancer. 2019. PMID: 31777602 Free PMC article.
-
A Four-Gene Promoter Methylation Marker Panel Consisting of GREM1, NEURL, LAD1, and NEFH Predicts Survival of Clear Cell Renal Cell Cancer Patients.Clin Cancer Res. 2017 Apr 15;23(8):2006-2018. doi: 10.1158/1078-0432.CCR-16-1236. Epub 2016 Oct 18. Clin Cancer Res. 2017. PMID: 27756787
Cited by
-
Focus on the Dynamics of Neutrophil-to-Lymphocyte Ratio in Cancer Patients Treated with Immune Checkpoint Inhibitors: A Meta-Analysis and Systematic Review.Cancers (Basel). 2022 Oct 27;14(21):5297. doi: 10.3390/cancers14215297. Cancers (Basel). 2022. PMID: 36358716 Free PMC article. Review.
-
SPI1 is a prognostic biomarker of immune infiltration and immunotherapy efficacy in clear cell renal cell carcinoma.Discov Oncol. 2022 Dec 7;13(1):134. doi: 10.1007/s12672-022-00592-0. Discov Oncol. 2022. PMID: 36477668 Free PMC article.
-
Epigenetic regulation and therapeutic targets in the tumor microenvironment.Mol Biomed. 2023 Jun 5;4(1):17. doi: 10.1186/s43556-023-00126-2. Mol Biomed. 2023. PMID: 37273004 Free PMC article. Review.
-
CD86 Molecule Might Be a Novel Immune-Related Prognostic Biomarker for Patients With Bladder Cancer by Bioinformatics and Experimental Assays.Front Oncol. 2021 Aug 6;11:679851. doi: 10.3389/fonc.2021.679851. eCollection 2021. Front Oncol. 2021. PMID: 34422632 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

