Development of a highly efficient 50K single nucleotide polymorphism genotyping array for the large and complex genome of Norway spruce (Picea abies L. Karst) by whole genome resequencing and its transferability to other spruce species
- PMID: 33179386
- PMCID: PMC7984398
- DOI: 10.1111/1755-0998.13292
Development of a highly efficient 50K single nucleotide polymorphism genotyping array for the large and complex genome of Norway spruce (Picea abies L. Karst) by whole genome resequencing and its transferability to other spruce species
Abstract
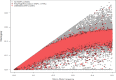
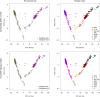
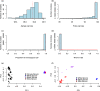
Norway spruce (Picea abies L. Karst) is one of the most important forest tree species with significant economic and ecological impact in Europe. For decades, genomic and genetic studies on Norway spruce have been challenging due to the large and repetitive genome (19.6 Gb with more than 70% being repetitive). To accelerate genomic studies, including population genetics, genome-wide association studies (GWAS) and genomic selection (GS), in Norway spruce and related species, we here report on the design and performance of a 50K single nucleotide polymorphism (SNP) genotyping array for Norway spruce. The array is developed based on whole genome resequencing (WGS), making it the first WGS-based SNP array in any conifer species so far. After identifying SNPs using genome resequencing data from 29 trees collected in northern Europe, we adopted a two-step approach to design the array. First, we built a 450K screening array and used this to genotype a population of 480 trees sampled from both natural and breeding populations across the Norway spruce distribution range. These samples were then used to select high-confidence probes that were put on the final 50K array. The SNPs selected are distributed over 45,552 scaffolds from the P. abies version 1.0 genome assembly and target 19,954 unique gene models with an even coverage of the 12 linkage groups in Norway spruce. We show that the array has a 99.5% probe specificity, >98% Mendelian allelic inheritance concordance, an average sample call rate of 96.30% and an SNP call rate of 98.90% in family trios and haploid tissues. We also observed that 23,797 probes (50%) could be identified with high confidence in three other spruce species (white spruce [Picea glauca], black spruce [P. mariana] and Sitka spruce [P. sitchensis]). The high-quality genotyping array will be a valuable resource for genetic and genomic studies in Norway spruce as well as in other conifer species of the same genus.
Keywords: Norway spruce; SNP array; genetic diversity; genome resequencing; genomic selection.
© 2020 The Authors. Molecular Ecology Resources published by John Wiley & Sons Ltd.
Figures



References
-
- Affymetrix (2016). Axiom analysis suite user guide (version 2.0). Affymetrix Inc.
-
- Azaiez, A. , Pavy, N. , Gérardi, S. , Laroche, J. , Boyle, B. , Gagnon, F. , Mottet, M.‐J. , Beaulieu, J. , & Bousquet, J. (2018). A catalog of annotated high‐confidence SNPs from exome capture and sequencing reveals highly polymorphic genes in Norway spruce (Picea abies). BMC Genomics, 19, 942. 10.1186/s12864-018-5247-z - DOI - PMC - PubMed
-
- Bianco, L. , Cestaro, A. , Linsmith, G. , Muranty, H. , Denancé, C. , Théron, A. , Poncet, C. , Micheletti, D. , Kerschbamer, E. , Di Pierro, E. A. , Larger, S. , Pindo, M. , Van de Weg, E. , Davassi, A. , Laurens, F. , Velasco, R. , Durel, C.‐E. , & Troggio, M. (2016). Development and validation of the Axiom®Apple480K SNP genotyping array. Plant J, 86, 62–74. 10.1111/tpj.13145 - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

