A rat epigenetic clock recapitulates phenotypic aging and co-localizes with heterochromatin
- PMID: 33179594
- PMCID: PMC7661040
- DOI: 10.7554/eLife.59201
A rat epigenetic clock recapitulates phenotypic aging and co-localizes with heterochromatin
Abstract
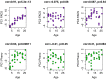
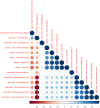
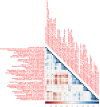
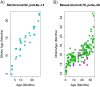
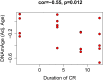
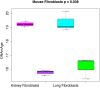
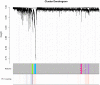
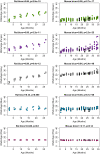
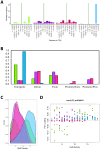
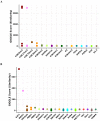
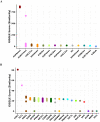
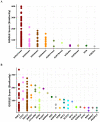
Robust biomarkers of aging have been developed from DNA methylation in humans and more recently, in mice. This study aimed to generate a novel epigenetic clock in rats-a model with unique physical, physiological, and biochemical advantages-by incorporating behavioral data, unsupervised machine learning, and network analysis to identify epigenetic signals that not only track with age, but also relates to phenotypic aging. Reduced representation bisulfite sequencing (RRBS) data was used to train an epigenetic age (DNAmAge) measure in Fischer 344 CDF (F344) rats. This measure correlated with age at (r = 0.93) in an independent sample, and related to physical functioning (p=5.9e-3), after adjusting for age and cell counts. DNAmAge was also found to correlate with age in male C57BL/6 mice (r = 0.79), and was decreased in response to caloric restriction. Our signatures driven by CpGs in intergenic regions that showed substantial overlap with H3K9me3, H3K27me3, and E2F1 transcriptional factor binding.
Keywords: DNA methylation; biological age; caloric restriction; computational biology; epigenetic clock; genetics; genomics; mouse; rat; systems biology.
Conflict of interest statement
ML, RM, MM, KP, AD, TM, CF, KT, MW, CD, MP, Rd, LF No competing interests declared
Figures



References
-
- Ambatipudi S, Horvath S, Perrier F, Cuenin C, Hernandez-Vargas H, Le Calvez-Kelm F, Durand G, Byrnes G, Ferrari P, Bouaoun L, Sklias A, Chajes V, Overvad K, Severi G, Baglietto L, Clavel-Chapelon F, Kaaks R, Barrdahl M, Boeing H, Trichopoulou A, Lagiou P, Naska A, Masala G, Agnoli C, Polidoro S, Tumino R, Panico S, Dollé M, Peeters PHM, Onland-Moret NC, Sandanger TM, Nøst TH, Weiderpass E, Quirós JR, Agudo A, Rodriguez-Barranco M, Huerta Castaño JM, Barricarte A, Fernández AM, Travis RC, Vineis P, Muller DC, Riboli E, Gunter M, Romieu I, Herceg Z. DNA methylome analysis identifies accelerated epigenetic ageing associated with postmenopausal breast Cancer susceptibility. European Journal of Cancer. 2017;75:299–307. doi: 10.1016/j.ejca.2017.01.014. - DOI - PMC - PubMed
-
- Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan JB, Gao Y, Deconde R, Chen M, Rajapakse I, Friend S, Ideker T, Zhang K. Genome-wide methylation profiles reveal quantitative views of human aging rates. Molecular Cell. 2013;49:359–367. doi: 10.1016/j.molcel.2012.10.016. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions

