Heterologous Expression of Cryptomaldamide in a Cyanobacterial Host
- PMID: 33180461
- PMCID: PMC8514081
- DOI: 10.1021/acssynbio.0c00431
Heterologous Expression of Cryptomaldamide in a Cyanobacterial Host
Abstract
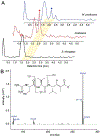
Filamentous marine cyanobacteria make a variety of bioactive molecules that are produced by polyketide synthases, nonribosomal peptide synthetases, and hybrid pathways that are encoded by large biosynthetic gene clusters. These cyanobacterial natural products represent potential drug leads; however, thorough pharmacological investigations have been impeded by the limited quantity of compound that is typically available from the native organisms. Additionally, investigations of the biosynthetic gene clusters and enzymatic pathways have been difficult due to the inability to conduct genetic manipulations in the native producers. Here we report a set of genetic tools for the heterologous expression of biosynthetic gene clusters in the cyanobacteria Synechococcus elongatus PCC 7942 and Anabaena (Nostoc) PCC 7120. To facilitate the transfer of gene clusters in both strains, we engineered a strain of Anabaena that contains S. elongatus homologous sequences for chromosomal recombination at a neutral site and devised a CRISPR-based strategy to efficiently obtain segregated double recombinant clones of Anabaena. These genetic tools were used to express the large 28.7 kb cryptomaldamide biosynthetic gene cluster from the marine cyanobacterium Moorena (Moorea) producens JHB in both model strains. S. elongatus did not produce cryptomaldamide; however, high-titer production of cryptomaldamide was obtained in Anabaena. The methods developed in this study will facilitate the heterologous expression of biosynthetic gene clusters isolated from marine cyanobacteria and complex metagenomic samples.
Keywords: cryptomaldamide; cyanobacteria; heterologous expression; natural products.
Conflict of interest statement
Conflict of Interest
W.H.G. has an equity interest in Sirenas Marine Discovery, Inc., a company that may potentially benefit from the research results, and also serves on the company’s Scientific Advisory Board. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies.
Figures


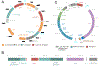

References
-
- Kleigrewe K; Gerwick L; Sherman DH; Gerwick WH, Unique marine derived cyanobacterial biosynthetic genes for chemical diversity. Nat Prod Rep 2016, 33 (2), 348–364. - PubMed
-
- Bullerjahn GS; McKay RM; Davis TW; Baker DB; Boyer GL; D'Anglada LV; Doucette GJ; Ho JC; Irwin EG; Kling CL; Kudela RM; Kurmayer R; Michalak AM; Ortiz JD; Otten TG; Paerl HW; Qin B; Sohngen BL; Stumpf RP; Visser PM; Wilhelm SW, Global solutions to regional problems: Collecting global expertise to address the problem of harmful cyanobacterial blooms. A Lake Erie case study. Harmful Algae 2016, 54, 223–238. - PMC - PubMed
-
- Dittmann E; Gugger M; Sivonen K; Fewer DP, Natural product biosynthetic diversity and comparative genomics of the cyanobacteria. Trends Microbiol 2015, 23 (10), 642–652. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

