Ikaros antagonizes DNA binding by STAT5 in pre-B cells
- PMID: 33180866
- PMCID: PMC7660478
- DOI: 10.1371/journal.pone.0242211
Ikaros antagonizes DNA binding by STAT5 in pre-B cells
Erratum in
-
Correction: Ikaros antagonizes DNA binding by STAT5 in pre-B cells.PLoS One. 2021 Jan 29;16(1):e0246570. doi: 10.1371/journal.pone.0246570. eCollection 2021. PLoS One. 2021. PMID: 33513189 Free PMC article.
Abstract
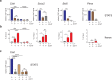
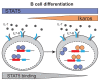
The IKZF1 gene, which encodes the Ikaros transcription factor, is frequently deleted or mutated in patients with B-cell precursor acute lymphoblastic leukemias that express oncogenes, like BCR-ABL, which activate the JAK-STAT5 pathway. Ikaros functionally antagonizes the transcriptional programs downstream of IL-7/STAT5 during B cell development, as well as STAT5 activity in leukemic cells. However, the mechanisms by which Ikaros interferes with STAT5 function is unknown. We studied the genomic distribution of Ikaros and STAT5 on chromatin in a murine pre-B cell line, and found that both proteins colocalize on >60% of STAT5 target regions. Strikingly, Ikaros activity leads to widespread loss of STAT5 binding at most of its genomic targets within two hours of Ikaros induction, suggesting a direct mechanism. Ikaros did not alter the level of total or phosphorylated STAT5 proteins, nor did it associate with STAT5. Using sequences from the Cish, Socs2 and Bcl6 genes that Ikaros and STAT5 target, we show that both proteins bind overlapping sequences at GGAA motifs. Our results demonstrate that Ikaros antagonizes STAT5 DNA binding, in part by competing for common target sequences. Our study has implications for understanding the functions of Ikaros and STAT5 in B cell development and transformation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Ikaros dominant negative isoform (Ik6) induces IL-3-independent survival of murine pro-B lymphocytes by activating JAK-STAT and up-regulating Bcl-xl levels.Leuk Lymphoma. 2008 May;49(5):965-73. doi: 10.1080/10428190801993462. Leuk Lymphoma. 2008. PMID: 18464116
-
Antagonism of B cell enhancer networks by STAT5 drives leukemia and poor patient survival.Nat Immunol. 2017 Jun;18(6):694-704. doi: 10.1038/ni.3716. Epub 2017 Apr 3. Nat Immunol. 2017. PMID: 28369050 Free PMC article.
-
Sleeping Beauty transposon screen identifies signaling modules that cooperate with STAT5 activation to induce B-cell acute lymphoblastic leukemia.Oncogene. 2016 Jun 30;35(26):3454-64. doi: 10.1038/onc.2015.405. Epub 2015 Oct 26. Oncogene. 2016. PMID: 26500062 Free PMC article.
-
Transcriptional circuits in B cell transformation.Curr Opin Hematol. 2017 Jul;24(4):345-352. doi: 10.1097/MOH.0000000000000352. Curr Opin Hematol. 2017. PMID: 28463873 Free PMC article. Review.
-
Ikaros fingers on lymphocyte differentiation.Int J Hematol. 2014 Sep;100(3):220-9. doi: 10.1007/s12185-014-1644-5. Epub 2014 Aug 2. Int J Hematol. 2014. PMID: 25085254 Free PMC article. Review.
Cited by
-
Ikaros deficiency is associated with aggressive BCR-ABL1 B-cell precursor acute lymphoblastic leukemia independent of the lineage and developmental origin.Haematologica. 2022 Jan 1;107(1):316-320. doi: 10.3324/haematol.2021.279125. Haematologica. 2022. PMID: 34587720 Free PMC article. No abstract available.
-
Age-associated myeloid malignancies - the role of STAT3 and STAT5 in myelodysplastic syndrome and acute myeloid leukemia.FEBS Lett. 2024 Nov;598(22):2809-2828. doi: 10.1002/1873-3468.14985. Epub 2024 Jul 24. FEBS Lett. 2024. PMID: 39048534 Free PMC article. Review.
-
The transcription factor Aiolos restrains the activation of intestinal intraepithelial lymphocytes.Nat Immunol. 2024 Jan;25(1):77-87. doi: 10.1038/s41590-023-01693-w. Epub 2023 Dec 4. Nat Immunol. 2024. PMID: 38049581 Free PMC article.
-
Aiolos represses CD4+ T cell cytotoxic programming via reciprocal regulation of TFH transcription factors and IL-2 sensitivity.Nat Commun. 2023 Mar 24;14(1):1652. doi: 10.1038/s41467-023-37420-0. Nat Commun. 2023. PMID: 36964178 Free PMC article.
-
Human influenza virus challenge identifies cellular correlates of protection for oral vaccination.Cell Host Microbe. 2021 Dec 8;29(12):1828-1837.e5. doi: 10.1016/j.chom.2021.10.009. Epub 2021 Nov 15. Cell Host Microbe. 2021. PMID: 34784508 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

