The genetic paradigms of dietary restriction fail to extend life span in cep-1(gk138) mutant of C. elegans p53 due to possible background mutations
- PMID: 33180887
- PMCID: PMC7660490
- DOI: 10.1371/journal.pone.0241478
The genetic paradigms of dietary restriction fail to extend life span in cep-1(gk138) mutant of C. elegans p53 due to possible background mutations
Abstract
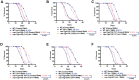
Dietary restriction (DR) increases life span and improves health in most model systems tested, including non-human primates. In C. elegans, as in other models, DR leads to reprogramming of metabolism, improvements in mitochondrial health, large changes in expression of cytoprotective genes and better proteostasis. Understandably, multiple global transcriptional regulators like transcription factors FOXO/DAF-16, FOXA/PHA-4, HSF1/HSF-1 and NRF2/SKN-1 are important for DR longevity. Considering the wide-ranging effects of p53 on organismal biology, we asked whether the C. elegans ortholog, CEP-1 is required for DR-mediated longevity assurance. We employed the widely-used TJ1 strain of cep-1(gk138). We show that cep-1(gk138) suppresses the life span extension of two genetic paradigms of DR, but two non-genetic modes of DR remain unaffected in this strain. We find that two aspects of DR, increased autophagy and up-regulation of the expression of cytoprotective xenobiotic detoxification program (cXDP) genes, are dampened in cep-1(gk138). Importantly, we find that background mutation(s) in the strain may be the actual cause for the phenotypic differences that we observed and cep-1 may not be directly involved in genetic DR-mediated longevity assurance in worms. Identifying these mutation(s) may reveal a novel regulator of longevity required specifically by genetic modes of DR.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Reduced expression of the Caenorhabditis elegans p53 ortholog cep-1 results in increased longevity.J Gerontol A Biol Sci Med Sci. 2007 Sep;62(9):951-9. doi: 10.1093/gerona/62.9.951. J Gerontol A Biol Sci Med Sci. 2007. PMID: 17895432
-
The effects of p53 on whole organism longevity are mediated by autophagy.Autophagy. 2008 Oct;4(7):870-3. doi: 10.4161/auto.6730. Epub 2008 Oct 4. Autophagy. 2008. PMID: 18728385
-
Pyruvate imbalance mediates metabolic reprogramming and mimics lifespan extension by dietary restriction in Caenorhabditis elegans.Aging Cell. 2011 Feb;10(1):39-54. doi: 10.1111/j.1474-9726.2010.00640.x. Epub 2010 Nov 15. Aging Cell. 2011. PMID: 21040400
-
Caenorhabditis elegans 2007: the premier model for the study of aging.Exp Gerontol. 2008 Jan;43(1):1-4. doi: 10.1016/j.exger.2007.09.008. Epub 2007 Oct 1. Exp Gerontol. 2008. PMID: 17977684 Free PMC article. Review.
-
DAF-16: FOXO in the Context of C. elegans.Curr Top Dev Biol. 2018;127:1-21. doi: 10.1016/bs.ctdb.2017.11.007. Epub 2018 Feb 2. Curr Top Dev Biol. 2018. PMID: 29433733 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

