WGVD: an integrated web-database for wheat genome variation and selective signatures
- PMID: 33181826
- PMCID: PMC7661095
- DOI: 10.1093/database/baaa090
WGVD: an integrated web-database for wheat genome variation and selective signatures
Abstract
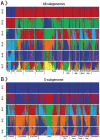
Bread wheat is one of the most important crops worldwide. With the release of the complete wheat reference genome and the development of next-generation sequencing technology, a mass of genomic data from bread wheat and its progenitors has been yield and has provided genomic resources for wheat genetics research. To conveniently and effectively access and use these data, we established Wheat Genome Variation Database, an integrated web-database including genomic variations from whole-genome resequencing and exome-capture data for bread wheat and its progenitors, as well as selective signatures during the process of wheat domestication and improvement. In this version, WGVD contains 7 346 814 single nucleotide polymorphisms (SNPs) and 1 044 400 indels focusing on genic regions and upstream or downstream regions. We provide allele frequency distribution patterns of these variations for 5 ploidy wheat groups or 17 worldwide bread wheat groups, the annotation of the variant types and the genotypes of all individuals for 2 versions of bread wheat reference genome (IWGSC RefSeq v1.0 and IWGSC RefSeq v2.0). Selective footprints for Aegilops tauschii, wild emmer, domesticated emmer, bread wheat landrace and bread wheat variety are evaluated with two statistical tests (FST and Pi) based on SNPs from whole-genome resequencing data. In addition, we provide the Genome Browser to visualize the genomic variations, the selective footprints, the genotype patterns and the read coverage depth, and the alignment tool Blast to search the homologous regions between sequences. All of these features of WGVD will promote wheat functional studies and wheat breeding.
Database url: http://animal.nwsuaf.edu.cn/code/index.php/Wheat.
© The Author(s) 2020. Published by Oxford University Press.
Figures






Similar articles
-
Frequent intra- and inter-species introgression shapes the landscape of genetic variation in bread wheat.Genome Biol. 2019 Jul 12;20(1):136. doi: 10.1186/s13059-019-1744-x. Genome Biol. 2019. PMID: 31300020 Free PMC article.
-
An online database for einkorn wheat to aid in gene discovery and functional genomics studies.Database (Oxford). 2023 Nov 16;2023:baad079. doi: 10.1093/database/baad079. Database (Oxford). 2023. PMID: 37971714 Free PMC article.
-
Seed-Derived Microbial Colonization of Wild Emmer and Domesticated Bread Wheat (Triticum dicoccoides and T. aestivum) Seedlings Shows Pronounced Differences in Overall Diversity and Composition.mBio. 2020 Nov 17;11(6):e02637-20. doi: 10.1128/mBio.02637-20. mBio. 2020. PMID: 33203759 Free PMC article.
-
Bread wheat: a role model for plant domestication and breeding.Hereditas. 2019 May 29;156:16. doi: 10.1186/s41065-019-0093-9. eCollection 2019. Hereditas. 2019. PMID: 31160891 Free PMC article. Review.
-
Deciphering the evolution and complexity of wheat germplasm from a genomic perspective.J Genet Genomics. 2023 Nov;50(11):846-860. doi: 10.1016/j.jgg.2023.08.002. Epub 2023 Aug 21. J Genet Genomics. 2023. PMID: 37611848 Review.
Cited by
-
Characterization of Expression and Epigenetic Features of Core Genes in Common Wheat.Genes (Basel). 2022 Jun 21;13(7):1112. doi: 10.3390/genes13071112. Genes (Basel). 2022. PMID: 35885895 Free PMC article.
-
Transcriptome Analysis of Roots from Wheat (Triticum aestivum L.) Varieties in Response to Drought Stress.Int J Mol Sci. 2023 Apr 14;24(8):7245. doi: 10.3390/ijms24087245. Int J Mol Sci. 2023. PMID: 37108408 Free PMC article.
-
Evolutionary Dynamics of Plant TRM6/TRM61 Complexes.Plants (Basel). 2025 Jun 11;14(12):1778. doi: 10.3390/plants14121778. Plants (Basel). 2025. PMID: 40573769 Free PMC article.
-
The Genomic Variation and Differentially Expressed Genes on the 6P Chromosomes in Wheat-Agropyron cristatum Addition Lines 5113 and II-30-5 Confer Different Desirable Traits.Int J Mol Sci. 2023 Apr 11;24(8):7056. doi: 10.3390/ijms24087056. Int J Mol Sci. 2023. PMID: 37108219 Free PMC article.
-
Wheat genomic study for genetic improvement of traits in China.Sci China Life Sci. 2022 Sep;65(9):1718-1775. doi: 10.1007/s11427-022-2178-7. Epub 2022 Aug 24. Sci China Life Sci. 2022. PMID: 36018491 Review.
References
-
- Pont C., Leroy T., Seidel M. et al. (2019) Tracing the ancestry of modern bread wheats. Nat. Genet., 51, 905–911. - PubMed
-
- Dvorak J., Luo M.-C., Yang Z.-L. et al. (1998) The structure of the Aegilops tauschii genepool and the evolution of hexaploid wheat. Theor. Appl. Genet., 97, 657–670.
-
- Luo M.-C., Yang Z.-L., You F.M. et al. (2007) The structure of wild and domesticated emmer wheat populations, gene flow between them, and the site of emmer domestication. Theor. Appl. Genet., 114, 947–959. - PubMed
-
- Lopes M.S., El-Basyoni I., Baenziger P.S. et al. (2015) Exploiting genetic diversity from landraces in wheat breeding for adaptation to climate change. J. Exp. Bot., 66, 3477–3486. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

