Plant Non-Coding RNAs: Origin, Biogenesis, Mode of Action and Their Roles in Abiotic Stress
- PMID: 33182372
- PMCID: PMC7664903
- DOI: 10.3390/ijms21218401
Plant Non-Coding RNAs: Origin, Biogenesis, Mode of Action and Their Roles in Abiotic Stress
Abstract
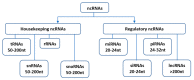
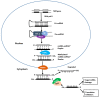
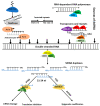
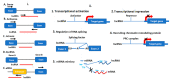
As sessile species, plants have to deal with the rapidly changing environment. In response to these environmental conditions, plants employ a plethora of response mechanisms that provide broad phenotypic plasticity to allow the fine-tuning of the external cues related reactions. Molecular biology has been transformed by the major breakthroughs in high-throughput transcriptome sequencing and expression analysis using next-generation sequencing (NGS) technologies. These innovations have provided substantial progress in the identification of genomic regions as well as underlying basis influencing transcriptional and post-transcriptional regulation of abiotic stress response. Non-coding RNAs (ncRNAs), particularly microRNAs (miRNAs), short interfering RNAs (siRNAs), and long non-coding RNAs (lncRNAs), have emerged as essential regulators of plants abiotic stress response. However, shared traits in the biogenesis of ncRNAs and the coordinated cross-talk among ncRNAs mechanisms contribute to the complexity of these molecules and might play an essential part in regulating stress responses. Herein, we highlight the current knowledge of plant microRNAs, siRNAs, and lncRNAs, focusing on their origin, biogenesis, modes of action, and fundamental roles in plant response to abiotic stresses.
Keywords: abiotic stress; biogenesis; long non-coding RNA; non-coding RNA; transcriptional.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

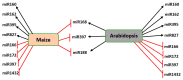
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

