Dimeric and Multimeric DNA Aptamers for Highly Effective Protein Recognition
- PMID: 33182593
- PMCID: PMC7698228
- DOI: 10.3390/molecules25225227
Dimeric and Multimeric DNA Aptamers for Highly Effective Protein Recognition
Abstract
Multivalent interactions frequently occur in biological systems and typically provide higher binding affinity and selectivity in target recognition than when only monovalent interactions are operative. Thus, taking inspiration by nature, bivalent or multivalent nucleic acid aptamers recognizing a specific biological target have been extensively studied in the last decades. Indeed, oligonucleotide-based aptamers are suitable building blocks for the development of highly efficient multivalent systems since they can be easily modified and assembled exploiting proper connecting linkers of different nature. Thus, substantial research efforts have been put in the construction of dimeric/multimeric versions of effective aptamers with various degrees of success in target binding affinity or therapeutic activity enhancement. The present review summarizes recent advances in the design and development of dimeric and multimeric DNA-based aptamers, including those forming G-quadruplex (G4) structures, recognizing different key proteins in relevant pathological processes. Most of the designed constructs have shown improved performance in terms of binding affinity or therapeutic activity as anti-inflammatory, antiviral, anticoagulant, and anticancer agents and their number is certainly bound to grow in the next future.
Keywords: G-quadruplex; aptamer; design; dimerization; molecular recognition; multivalency; protein target; therapy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


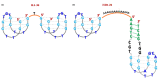
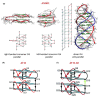
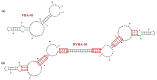
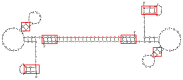




Similar articles
-
G-quadruplex-based aptamers against protein targets in therapy and diagnostics.Biochim Biophys Acta Gen Subj. 2017 May;1861(5 Pt B):1429-1447. doi: 10.1016/j.bbagen.2016.11.027. Epub 2016 Nov 16. Biochim Biophys Acta Gen Subj. 2017. PMID: 27865995 Free PMC article. Review.
-
Nucleic acid aptamers based on the G-quadruplex structure: therapeutic and diagnostic potential.Curr Med Chem. 2009;16(10):1248-65. doi: 10.2174/092986709787846640. Curr Med Chem. 2009. PMID: 19355883 Review.
-
Tuning the Polymorphism of the Anti-VEGF G-rich V7t1 Aptamer by Covalent Dimeric Constructs.Int J Mol Sci. 2020 Mar 13;21(6):1963. doi: 10.3390/ijms21061963. Int J Mol Sci. 2020. PMID: 32183039 Free PMC article.
-
Multifunctional G-quadruplex aptamers and their application to protein detection.Chemistry. 2009;15(4):1036-42. doi: 10.1002/chem.200801282. Chemistry. 2009. PMID: 19053089
-
Selective light-up of dimeric G-quadruplex forming aptamers for efficient VEGF165 detection.Int J Biol Macromol. 2023 Jan 1;224:344-357. doi: 10.1016/j.ijbiomac.2022.10.128. Epub 2022 Oct 19. Int J Biol Macromol. 2023. PMID: 36270405
Cited by
-
Covalent Bi-Modular Parallel and Antiparallel G-Quadruplex DNA Nanocostructs Reduce Viability of Patient Glioma Primary Cell Cultures.Int J Mol Sci. 2021 Mar 25;22(7):3372. doi: 10.3390/ijms22073372. Int J Mol Sci. 2021. PMID: 33806042 Free PMC article.
-
Three on Three: Universal and High-Affinity Molecular Recognition of the Symmetric Homotrimeric Spike Protein of SARS-CoV-2 with a Symmetric Homotrimeric Aptamer.J Am Chem Soc. 2022 Dec 28;144(51):23465-23473. doi: 10.1021/jacs.2c09870. Epub 2022 Dec 15. J Am Chem Soc. 2022. PMID: 36520671 Free PMC article.
-
Affinity Chromatography-Based Assays for the Screening of Potential Ligands Selective for G-Quadruplex Structures.ChemistryOpen. 2022 May;11(5):e202200090. doi: 10.1002/open.202200090. ChemistryOpen. 2022. PMID: 35608081 Free PMC article.
-
High-Affinity Dimeric Aptamers Enable the Rapid Electrochemical Detection of Wild-Type and B.1.1.7 SARS-CoV-2 in Unprocessed Saliva.Angew Chem Int Ed Engl. 2021 Nov 2;60(45):24266-24274. doi: 10.1002/anie.202110819. Epub 2021 Oct 4. Angew Chem Int Ed Engl. 2021. PMID: 34464491 Free PMC article.
-
Genetically Encoded Fluorogenic DNA Aptamers for Imaging Metabolite in Living Cells.J Am Chem Soc. 2025 Jan 15;147(2):1529-1541. doi: 10.1021/jacs.4c09855. Epub 2024 Dec 31. J Am Chem Soc. 2025. PMID: 39739942 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

