Natural Products Attenuating Biosynthesis, Processing, and Activity of Ras Oncoproteins: State of the Art and Future Perspectives
- PMID: 33182807
- PMCID: PMC7698260
- DOI: 10.3390/biom10111535
Natural Products Attenuating Biosynthesis, Processing, and Activity of Ras Oncoproteins: State of the Art and Future Perspectives
Abstract
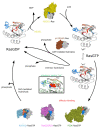
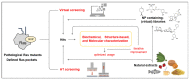
RAS genes encode signaling proteins, which, in mammalian cells, act as molecular switches regulating critical cellular processes as proliferation, growth, differentiation, survival, motility, and metabolism in response to specific stimuli. Deregulation of Ras functions has a high impact on human health: gain-of-function point mutations in RAS genes are found in some developmental disorders and thirty percent of all human cancers, including the deadliest. For this reason, the pathogenic Ras variants represent important clinical targets against which to develop novel, effective, and possibly selective pharmacological inhibitors. Natural products represent a virtually unlimited resource of structurally different compounds from which one could draw on for this purpose, given the improvements in isolation and screening of active molecules from complex sources. After a summary of Ras proteins molecular and regulatory features and Ras-dependent pathways relevant for drug development, we point out the most promising inhibitory approaches, the known druggable sites of wild-type and oncogenic Ras mutants, and describe the known natural compounds capable of attenuating Ras signaling. Finally, we highlight critical issues and perspectives for the future selection of potential Ras inhibitors from natural sources.
Keywords: Ras druggable pockets; Ras inhibitors; Ras inhibitory strategies; Ras oncogenes; Ras signaling; anticarcinogenic effect; cancer; natural products.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

