Protein complexes and neighborhoods driving autophagy
- PMID: 33183148
- PMCID: PMC8526019
- DOI: 10.1080/15548627.2020.1847461
Protein complexes and neighborhoods driving autophagy
Abstract
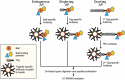
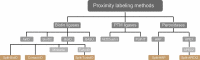
Autophagy summarizes evolutionarily conserved, intracellular degradation processes targeting cytoplasmic material for lysosomal degradation. These encompass constitutive processes as well as stress responses, which are often found dysregulated in diseases. Autophagy pathways help in the clearance of damaged organelles, protein aggregates and macromolecules, mediating their recycling and maintaining cellular homeostasis. Protein-protein interaction networks contribute to autophagosome biogenesis, substrate loading, vesicular trafficking and fusion, protein translocations across membranes and degradation in lysosomes. Hypothesis-free proteomic approaches tremendously helped in the functional characterization of protein-protein interactions to uncover molecular mechanisms regulating autophagy. In this review, we elaborate on the importance of understanding protein-protein-interactions of varying affinities and on the strengths of mass spectrometry-based proteomic approaches to study these, generating new mechanistic insights into autophagy regulation. We discuss in detail affinity purification approaches and recent developments in proximity labeling coupled to mass spectrometry, which uncovered molecular principles of autophagy mechanisms.Abbreviations: AMPK: AMP-activated protein kinase; AP-MS: affinity purification-mass spectrometry; APEX2: ascorbate peroxidase-2; ATG: autophagy related; BioID: proximity-dependent biotin identification; ER: endoplasmic reticulum; GFP: green fluorescent protein; iTRAQ: isobaric tag for relative and absolute quantification; MS: mass spectrometry; PCA: protein-fragment complementation assay; PL-MS: proximity labeling-mass spectrometry; PtdIns3P: phosphatidylinositol-3-phosphate; PTM: posttranslational modification; PUP-IT: pupylation-based interaction tagging; RFP: red fluorescent protein; SILAC: stable isotope labeling by amino acids in cell culture; TAP: tandem affinity purification; TMT: tandem mass tag.
Keywords: Autophagy; affinity purification; mass spectrometry; protein-protein interactions; proximity labeling; quantitative proteomics.
Conflict of interest statement
The authors declare no competing interests
Figures



References
-
- Mizushima N, Klionsky DJ.. Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr. 2007;27:19–40. - PubMed
-
- Chiang HL, Terlecky SR, Plant CP, et al. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science. 1989. Oct 20;246(4928):382–385. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
