Identification of natural inhibitors against prime targets of SARS-CoV-2 using molecular docking, molecular dynamics simulation and MM-PBSA approaches
- PMID: 33183178
- PMCID: PMC7678369
- DOI: 10.1080/07391102.2020.1846624
Identification of natural inhibitors against prime targets of SARS-CoV-2 using molecular docking, molecular dynamics simulation and MM-PBSA approaches
Abstract
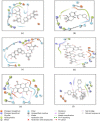
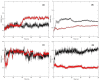
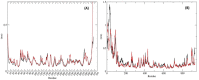
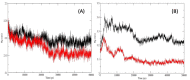
The recently emerged COVID-19 has been declared a pandemic by the World Health Organization as to date; no therapeutic drug/vaccine is available for the treatment. Due to the lack of time and the urgency to contain the pandemic, computational screening appears to be the best tool to find a therapeutic solution. Accumulated evidence suggests that many phyto-compounds possess anti-viral activity. Therefore, we identified possible phyto-compounds that could be developed and used for COVID-19 treatment. In particular, molecular docking was used to prioritize the possible active phyto-compounds against two key targets namely RNA dependent RNA polymerase (RdRp) and main protease (Mpro) of SARS-CoV-2. In this study, an antiviral drug- Remdesivir (RdRp inhibitor) and Darunavir (Mpro inhibitor) are used as reference drugs. This study revealed that phyto-molecules- Mulberroside-A/C/E/F, Emblicanin A, Nimbolide, and Punigluconin showed high binding affinity against RdRp while Andrographolides, Mulberrosides, Anolignans, Chebulic acid, Mimusopic acid, and Punigluconin showed better binding affinity against Mpro as compared with the reference drug. Furthermore, ADME profiles validated the drug-likeness properties of prioritized phyto-compounds. Besides, to assess the stability, MD simulations studies were performed along with reference inhibitors for Mpro (Darunavir) and RdRp (Remdesivir). Binding free energy calculations (MM-PBSA) revealed the estimated value (ΔG) of Mpro_Darunavir; Mpro_Mulberroside E; RdRp_Remdesivir and RdRp_Emblicanin A were -111.62 ± 6.788, -141.443 ± 9.313, 30.782 ± 5.85 and -89.424 ± 3.130 kJmol-1, respectively. Taken together, the study revealed the potential of these phyto-compounds as inhibitors of RdRp and Mpro inhibitor that could be further validated against SARS-CoV-2 for clinical benefits.Communicated by Ramaswamy H. Sarma.
Keywords: COVID-19; MM-PBSA; SARS-CoV-2; molecular docking; molecular dynamics; phyto-compounds.
Conflict of interest statement
There are no conflicts of interest.
Figures

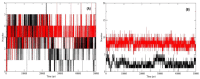


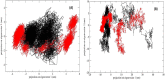
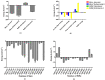
References
-
- Berendsen, H. J. C., Postma, J. P. M., Van Gunsteren, W. F., Dinola, A., & Haak, J. R. (1984). Molecular dynamics with coupling to an external bath. The Journal of Chemical Physics, 81(8), 3684–3690. 10.1063/1.448118 - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
