Microglial responses to peripheral type 1 interferon
- PMID: 33183319
- PMCID: PMC7659169
- DOI: 10.1186/s12974-020-02003-z
Microglial responses to peripheral type 1 interferon
Abstract
Background: Interferon α (IFNα) is a cytokine whose production is increased endogenously in response to viral infection and in autoimmune diseases such as systemic lupus erythematosus (SLE). An elevated IFNα signature has been associated with clinically observed neuro-behavioural deficits such as mild cognitive impairment, fatigue, depression and psychosis in these diseases. However, the mechanisms underlying these neuropsychiatric symptoms remain largely unknown, and it is as yet unclear how IFNα signalling might influence central nervous system (CNS) function. Aberrant microglia-mediated synaptic pruning and function has recently been implicated in several neurodegenerative and neuropsychiatric diseases, but whether and how IFNα modulates these functions are not well defined.
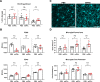
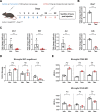
Methods: Using a model of peripheral IFNα administration, we investigated gene expression changes due to IFNAR signalling in microglia. Bulk RNA sequencing on sorted microglia from wild type and microglia-specific Ifnar1 conditional knockout mice was performed to evaluate IFNα and IFNAR signalling-dependent changes in gene expression. Furthermore, the effects of IFNα on microglia morphology and synapse engulfment were assessed, via immunohistochemistry and flow cytometry.
Results: We found that IFNα exposure through the periphery induces a unique gene signature in microglia that includes the expected upregulation of multiple interferon-stimulated genes (ISGs), as well as the complement component C4b. We additionally characterized several IFNα-dependent changes in microglial phenotype, including expression of CD45 and CD68, cellular morphology and presynaptic engulfment, that reveal subtle brain region-specific differences. Finally, by specifically knocking down expression of IFNAR1 on microglia, we show that these changes are largely attributable to direct IFNAR signalling on microglia and not from indirect signalling effects through other CNS parenchymal cell types which are capable of IFNα-IFNAR signal transduction.
Conclusions: Peripheral IFNα induces unique genetic and phenotypic changes in microglia that are largely dependent on direct signalling through microglial IFNAR. The IFNα-induced upregulation of C4b could play important roles in the context of aberrant synaptic pruning in neuropsychiatric disease.
Keywords: Complement; Interferon alpha (IFNα); Interferon-stimulated gene (ISG); Microglia; Neuropsychiatric; Synapse engulfment.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

