Estrogen Receptor Alpha Mutations in Breast Cancer Cells Cause Gene Expression Changes through Constant Activity and Secondary Effects
- PMID: 33184109
- PMCID: PMC7854489
- DOI: 10.1158/0008-5472.CAN-20-1171
Estrogen Receptor Alpha Mutations in Breast Cancer Cells Cause Gene Expression Changes through Constant Activity and Secondary Effects
Abstract
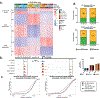
While breast cancer patients with tumors that express estrogen receptor α (ER) generally respond well to hormone therapies that block ER activity, a significant number of patients relapse. Approximately 30% of these recurrences harbor activating mutations in the ligand binding domain (LBD) of ER, which have been shown to confer ligand-independent function. However, much is still unclear regarding the effect of mutant ER beyond its estrogen independence. To investigate the molecular effects of mutant ER, we developed multiple isogenic ER-mutant cell lines for the most common LBD mutations, Y537S and D538G. These mutations induced differential expression of thousands of genes, the majority of which were mutant allele specific and were not observed upon estrogen treatment of wild-type (WT) cells. These mutant-specific genes showed consistent differential expression across ER-mutant lines developed in other laboratories. WT cells with long-term estrogen exposure only exhibited some of these transcriptional changes, suggesting that mutant ER causes novel regulatory effects that are not simply due to constant activity. While ER mutations exhibited minor effects on ER genomic binding, with the exception of ligand independence, ER mutations conferred substantial differences in chromatin accessibility. Mutant ER was bound to approximately a quarter of mutant-enriched accessible regions that were enriched for other DNA binding factors, including FOXA1, CTCF, and OCT1. Overall, our findings indicate that mutant ER causes several consistent effects on gene expression, both indirectly and through constant activity. SIGNIFICANCE: This study demonstrates the multiple roles of mutant ER in breast cancer progression, including constant ER activity and secondary regulatory effects on gene expression and chromatin accessibility. GRAPHICAL ABSTRACT: http://cancerres.aacrjournals.org/content/canres/81/3/539/F1.large.jpg.See related commentary by Hermida-Prado and Jeselsohn, p. 537 See related article by Williams and colleagues, p. 732.
©2020 American Association for Cancer Research.
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures





Comment in
-
Steroid Hormone Receptor and Infiltrating Immune Cell Status Reveals Therapeutic Vulnerabilities of ESR1-Mutant Breast Cancer.Cancer Res. 2021 Feb 1;81(3):732-746. doi: 10.1158/0008-5472.CAN-20-1200. Epub 2020 Nov 12. Cancer Res. 2021. PMID: 33184106 Free PMC article.
-
The ESR1 Mutations: From Bedside to Bench to Bedside.Cancer Res. 2021 Feb 1;81(3):537-538. doi: 10.1158/0008-5472.CAN-20-4037. Cancer Res. 2021. PMID: 33526469
Comment on
-
The ESR1 Mutations: From Bedside to Bench to Bedside.Cancer Res. 2021 Feb 1;81(3):537-538. doi: 10.1158/0008-5472.CAN-20-4037. Cancer Res. 2021. PMID: 33526469
References
-
- Blows FM, Driver KE, Schmidt MK, Broeks A, van Leeuwen FE, Wesseling J, et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: a collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med 2010;7:e1000279. - PMC - PubMed
-
- Group EBCTC. Tamoxifen for early breast cancer: an overview of the randomised trials. Lancet 1998;351:1451–67 - PubMed
-
- Group EBCTC. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet 2005;365:1687–717 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

