A ubiquitin ligase mediates target-directed microRNA decay independently of tailing and trimming
- PMID: 33184234
- PMCID: PMC8177725
- DOI: 10.1126/science.abc9546
A ubiquitin ligase mediates target-directed microRNA decay independently of tailing and trimming
Abstract
MicroRNAs (miRNAs) act in concert with Argonaute (AGO) proteins to repress target messenger RNAs. After AGO loading, miRNAs generally exhibit slow turnover. An important exception occurs when miRNAs encounter highly complementary targets, which can trigger a process called target-directed miRNA degradation (TDMD). During TDMD, miRNAs undergo tailing and trimming, suggesting that this is an important step in the decay mechanism. We identified a cullin-RING ubiquitin ligase (CRL), containing the substrate adaptor ZSWIM8, that mediates TDMD. The ZSWIM8 CRL interacts with AGO proteins, promotes TDMD in a tailing and trimming-independent manner, and regulates miRNA expression in multiple cell types. These findings suggest a model in which the ZSWIM8 ubiquitin ligase mediates TDMD by directing proteasomal decay of miRNA-containing complexes engaged with highly complementary targets.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures



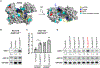

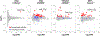
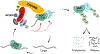
Comment in
-
To Degrade a MicroRNA, Destroy Its Argonaute Protein.Mol Cell. 2021 Jan 21;81(2):223-225. doi: 10.1016/j.molcel.2020.12.043. Mol Cell. 2021. PMID: 33482091
References
-
- Treiber T, Treiber N, Meister G, Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol 20, 5–20 (2019). - PubMed
-
- Hwang HW, Wentzel EA, Mendell JT, A hexanucleotide element directs microRNA nuclear import. Science 315, 97–100 (2007). - PubMed
-
- Krol J et al., Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell 141, 618–631 (2010). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

