Engineering of α-PD-1 antibody-expressing long-lived plasma cells by CRISPR/Cas9-mediated targeted gene integration
- PMID: 33184267
- PMCID: PMC7661525
- DOI: 10.1038/s41419-020-03187-1
Engineering of α-PD-1 antibody-expressing long-lived plasma cells by CRISPR/Cas9-mediated targeted gene integration
Abstract
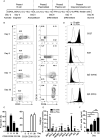
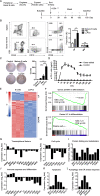
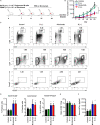
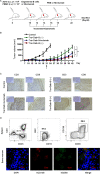
Long-lived plasma cells (LLPCs) are robust specialized antibody-secreting cells that mainly stay in the bone marrow and can persist a lifetime. As they can be generated by inducing the differentiation of B-lymphocytes, we investigated the possibility that human LLPCs might be engineered to express α-PD-1 monoclonal antibody to substitute recombinant α-PD-1 antitumor immunotherapy. To this end, we inserted an α-PD-1 cassette into the GAPDH locus through Cas9/sgRNA-guided specific integration in B-lymphocytes, which was mediated by an integrase-defective lentiviral vector. The edited B cells were capable of differentiating into LLPCs both in vitro and in vivo. Transcriptional profiling analysis confirmed that these cells were typical LLPCs. Importantly, these cells secreted de novo antibodies persistently, which were able to inhibit human melanoma growth via an antibody-mediated checkpoint blockade in xenograft-tumor mice. Our work suggests that the engineered LLPCs may be utilized as a vehicle to constantly produce special antibodies for long-term cellular immunotherapy to eradicate tumors and cellular reservoirs for various pathogens including human immunodeficiency virus type 1 (HIV-1) and hepatitis B virus (HBV).
Conflict of interest statement
The authors have declared that they have no conflict of interest.
Figures



Similar articles
-
Cas9-edited immune checkpoint blockade PD-1 DNA polyaptamer hydrogel for cancer immunotherapy.Biomaterials. 2019 Oct;218:119359. doi: 10.1016/j.biomaterials.2019.119359. Epub 2019 Jul 15. Biomaterials. 2019. PMID: 31349094
-
Nucleofection with Plasmid DNA for CRISPR/Cas9-Mediated Inactivation of Programmed Cell Death Protein 1 in CD133-Specific CAR T Cells.Hum Gene Ther. 2019 Apr;30(4):446-458. doi: 10.1089/hum.2017.234. Epub 2018 Apr 27. Hum Gene Ther. 2019. PMID: 29706119
-
Effect of CRISPR/Cas9-Mediated PD-1-Disrupted Primary Human Third-Generation CAR-T Cells Targeting EGFRvIII on In Vitro Human Glioblastoma Cell Growth.Cells. 2020 Apr 16;9(4):998. doi: 10.3390/cells9040998. Cells. 2020. PMID: 32316275 Free PMC article.
-
Factors That Govern the Induction of Long-Lived Antibody Responses.Viruses. 2020 Jan 7;12(1):74. doi: 10.3390/v12010074. Viruses. 2020. PMID: 31936163 Free PMC article. Review.
-
Long-Lived Plasma Cells in Mice and Men.Front Immunol. 2018 Nov 16;9:2673. doi: 10.3389/fimmu.2018.02673. eCollection 2018. Front Immunol. 2018. PMID: 30505309 Free PMC article. Review.
Cited by
-
Nanoliposomes as nonviral vectors in cancer gene therapy.MedComm (2020). 2024 Jun 25;5(7):e583. doi: 10.1002/mco2.583. eCollection 2024 Jul. MedComm (2020). 2024. PMID: 38919334 Free PMC article. Review.
-
Cell-specific delivery of CRISPR-Cas9 with pseudotyped lentiviral particles: Just change the envelope.Mol Ther Nucleic Acids. 2024 Nov 27;35(4):102395. doi: 10.1016/j.omtn.2024.102395. eCollection 2024 Dec 10. Mol Ther Nucleic Acids. 2024. PMID: 39669702 Free PMC article. No abstract available.
-
Human plasma cells engineered to secrete bispecifics drive effective in vivo leukemia killing.Mol Ther. 2024 Aug 7;32(8):2676-2691. doi: 10.1016/j.ymthe.2024.06.004. Epub 2024 Jul 2. Mol Ther. 2024. PMID: 38959896 Free PMC article.
-
Effect of CRISPR/Cas9-Edited PD-1/PD-L1 on Tumor Immunity and Immunotherapy.Front Immunol. 2022 Mar 1;13:848327. doi: 10.3389/fimmu.2022.848327. eCollection 2022. Front Immunol. 2022. PMID: 35300341 Free PMC article. Review.
-
Advancing in vivo genome editing: B cell engineering via adenoviral delivery systems.Mol Ther. 2023 Sep 6;31(9):2554-2556. doi: 10.1016/j.ymthe.2023.08.012. Epub 2023 Aug 23. Mol Ther. 2023. PMID: 37611582 Free PMC article. No abstract available.
References
-
- Melo ME, et al. Gene transfer of Ig-fusion proteins into B cells prevents and treats autoimmune diseases. J. Immunol. 2002;168:4788–4795. - PubMed
-
- Tellier J, Nutt SL. Plasma cells: the programming of an antibody-secreting machine. Eur. J. Immunol. 2019;49:30–37. - PubMed
-
- Nutt SL, Hodgkin PD, Tarlinton DM, Corcoran LM. The generation of antibody-secreting plasma cells. Nat. Rev. Immunol. 2015;15:160–171. - PubMed
-
- Kometani K, Kurosaki T. Differentiation and maintenance of long-lived plasma cells. Curr. Opin. Immunol. 2015;33:64–69. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

