Unraveling response to temozolomide in preclinical GL261 glioblastoma with MRI/MRSI using radiomics and signal source extraction
- PMID: 33184423
- PMCID: PMC7661707
- DOI: 10.1038/s41598-020-76686-y
Unraveling response to temozolomide in preclinical GL261 glioblastoma with MRI/MRSI using radiomics and signal source extraction
Abstract
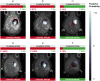
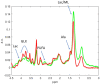
Glioblastoma is the most frequent aggressive primary brain tumor amongst human adults. Its standard treatment involves chemotherapy, for which the drug temozolomide is a common choice. These are heterogeneous and variable tumors which might benefit from personalized, data-based therapy strategies, and for which there is room for improvement in therapy response follow-up, investigated with preclinical models. This study addresses a preclinical question that involves distinguishing between treated and control (untreated) mice bearing glioblastoma, using machine learning techniques, from magnetic resonance-based data in two modalities: MRI and MRSI. It aims to go beyond the comparison of methods for such discrimination to provide an analytical pipeline that could be used in subsequent human studies. This analytical pipeline is meant to be a usable and interpretable tool for the radiology expert in the hope that such interpretation helps revealing new insights about the problem itself. For that, we propose coupling source extraction-based and radiomics-based data transformations with feature selection. Special attention is paid to the generation of radiologist-friendly visual nosological representations of the analyzed tumors.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

