Functional impacts of the ubiquitin-proteasome system on DNA damage recognition in global genome nucleotide excision repair
- PMID: 33184426
- PMCID: PMC7665181
- DOI: 10.1038/s41598-020-76898-2
Functional impacts of the ubiquitin-proteasome system on DNA damage recognition in global genome nucleotide excision repair
Abstract
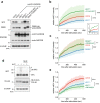
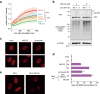
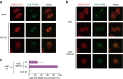
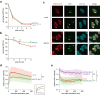
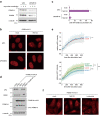
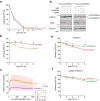
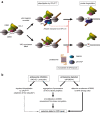
The ubiquitin-proteasome system (UPS) plays crucial roles in regulation of various biological processes, including DNA repair. In mammalian global genome nucleotide excision repair (GG-NER), activation of the DDB2-associated ubiquitin ligase upon UV-induced DNA damage is necessary for efficient recognition of lesions. To date, however, the precise roles of UPS in GG-NER remain incompletely understood. Here, we show that the proteasome subunit PSMD14 and the UPS shuttle factor RAD23B can be recruited to sites with UV-induced photolesions even in the absence of XPC, suggesting that proteolysis occurs at DNA damage sites. Unexpectedly, sustained inhibition of proteasome activity results in aggregation of PSMD14 (presumably with other proteasome components) at the periphery of nucleoli, by which DDB2 is immobilized and sequestered from its lesion recognition functions. Although depletion of PSMD14 alleviates such DDB2 immobilization induced by proteasome inhibitors, recruitment of DDB2 to DNA damage sites is then severely compromised in the absence of PSMD14. Because all of these proteasome dysfunctions selectively impair removal of cyclobutane pyrimidine dimers, but not (6-4) photoproducts, our results indicate that the functional integrity of the proteasome is essential for the DDB2-mediated lesion recognition sub-pathway, but not for GG-NER initiated through direct lesion recognition by XPC.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Cullin 4A-mediated proteolysis of DDB2 protein at DNA damage sites regulates in vivo lesion recognition by XPC.J Biol Chem. 2006 May 12;281(19):13404-13411. doi: 10.1074/jbc.M511834200. Epub 2006 Mar 8. J Biol Chem. 2006. PMID: 16527807
-
Mechanism and regulation of DNA damage recognition in mammalian nucleotide excision repair.Enzymes. 2019;45:99-138. doi: 10.1016/bs.enz.2019.06.004. Epub 2019 Jul 8. Enzymes. 2019. PMID: 31627884 Review.
-
Timely upstream events regulating nucleotide excision repair by ubiquitin-proteasome system: ubiquitin guides the way.DNA Repair (Amst). 2021 Jul;103:103128. doi: 10.1016/j.dnarep.2021.103128. Epub 2021 May 12. DNA Repair (Amst). 2021. PMID: 33991872 Free PMC article. Review.
-
UV radiation-induced SUMOylation of DDB2 regulates nucleotide excision repair.Carcinogenesis. 2017 Oct 1;38(10):976-985. doi: 10.1093/carcin/bgx076. Carcinogenesis. 2017. PMID: 28981631 Free PMC article.
-
Poly(ADP-ribose) polymerase 1 escorts XPC to UV-induced DNA lesions during nucleotide excision repair.Proc Natl Acad Sci U S A. 2017 Aug 15;114(33):E6847-E6856. doi: 10.1073/pnas.1706981114. Epub 2017 Jul 31. Proc Natl Acad Sci U S A. 2017. PMID: 28760956 Free PMC article.
Cited by
-
Exposure of cells to near-infrared irradiation relaxes chromatin compaction and facilitates recognition of cyclo-butane pyrimidine dimers.Sci Rep. 2025 Jul 1;15(1):22312. doi: 10.1038/s41598-025-08763-z. Sci Rep. 2025. PMID: 40594866 Free PMC article.
-
A Whole New Comprehension about ncRNA-Encoded Peptides/Proteins in Cancers.Cancers (Basel). 2022 Oct 23;14(21):5196. doi: 10.3390/cancers14215196. Cancers (Basel). 2022. PMID: 36358616 Free PMC article. Review.
-
Histone deacetylation regulates nucleotide excision repair through an interaction with the XPC protein.iScience. 2022 Mar 9;25(4):104040. doi: 10.1016/j.isci.2022.104040. eCollection 2022 Apr 15. iScience. 2022. PMID: 35330687 Free PMC article.
-
Chaperones for dancing on chromatin: Role of post-translational modifications in dynamic damage detection hand-offs during nucleotide excision repair.Bioessays. 2021 May;43(5):e2100011. doi: 10.1002/bies.202100011. Epub 2021 Feb 23. Bioessays. 2021. PMID: 33620094 Free PMC article.
-
Lesion recognition by XPC, TFIIH and XPA in DNA excision repair.Nature. 2023 May;617(7959):170-175. doi: 10.1038/s41586-023-05959-z. Epub 2023 Apr 19. Nature. 2023. PMID: 37076618 Free PMC article.
References
-
- Nishigori C, Sugasawa K, editors. DNA Repair Disorders. New York: Springer Nature; 2019.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

