Genetic influences on viral-induced cytokine responses in the lung
- PMID: 33184476
- PMCID: PMC7658619
- DOI: 10.1038/s41385-020-00355-6
Genetic influences on viral-induced cytokine responses in the lung
Abstract
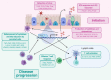
Infection with respiratory viruses such as influenza, respiratory syncytial virus and coronavirus provides a difficult immunological challenge for the host, where a balance must be established between controlling viral replication and limiting damage to the delicate lung structure. Although the genetic architecture of host responses to respiratory viral infections is not yet understood, it is clear there is underlying heritability that influences pathogenesis. Immune control of virus replication is essential in respiratory infections, but overt activation can enhance inflammation and disease severity. Cytokines initiate antiviral immune responses but are implicated in viral pathogenesis. Here, we discuss how host genetic variation may influence cytokine responses to respiratory viral infections and, based on our current understanding of the role that cytokines play in viral pathogenesis, how this may influence disease severity. We also discuss how induced pluripotent stem cells may be utilised to probe the mechanistic implications of allelic variation in genes in virus-induced inflammatory responses. Ultimately, this could help to design better immune modulators, stratify high risk patients and tailor anti-inflammatory treatments, potentially expanding the ability to treat respiratory virus outbreaks in the future.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

