Bacterial OTU deubiquitinases regulate substrate ubiquitination upon Legionella infection
- PMID: 33185526
- PMCID: PMC7690952
- DOI: 10.7554/eLife.58277
Bacterial OTU deubiquitinases regulate substrate ubiquitination upon Legionella infection
Abstract
Legionella pneumophila causes a severe pneumonia known as Legionnaires' disease. During the infection, Legionella injects more than 300 effector proteins into host cells. Among them are enzymes involved in altering the host-ubiquitination system. Here, we identified two
Keywords: Legionella pneumophila; OTU-deubiquitinase; bacterial deubiquitinase; biochemistry; chemical biology; effector proteins; human; ubiquitin.
© 2020, Shin et al.
Conflict of interest statement
DS, AB, YC, MA, AM, Gv, HO, GH No competing interests declared, ID Reviewing editor, eLife
Figures
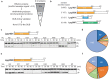
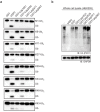





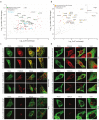
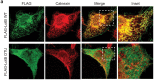

References
-
- Abdul Rehman SA, Kristariyanto YA, Choi SY, Nkosi PJ, Weidlich S, Labib K, Hofmann K, Kulathu Y. MINDY-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Molecular Cell. 2016;63:146–155. doi: 10.1016/j.molcel.2016.05.009. - DOI - PMC - PubMed
-
- Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1-2:19–25. doi: 10.1016/j.softx.2015.06.001. - DOI
-
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallographica Section D Biological Crystallography. 2012;68:352–367. doi: 10.1107/S0907444912001308. - DOI - PMC - PubMed
-
- Best RB, Zhu X, Shim J, Lopes PE, Mittal J, Feig M, Mackerell AD. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. Journal of Chemical Theory and Computation. 2012;8:3257–3273. doi: 10.1021/ct300400x. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- SFB1177/Deutsche Forschungsgemeinschaft/International
- CEF-MC-EXC115/2/Deutsche Forschungsgemeinschaft/International
- SFB902/Deutsche Forschungsgemeinschaft/International
- ID 259139777-SFB1177/Deutsche Forschungsgemeinschaft/International
- VI.Vidi.192.011/NWO/International
- 451001026/ZONMW_/ZonMw/Netherlands
- Project-ID III L6-519/03/03.001 -(0006)/LOEWE program DynaMem of the State of Hesse/International
- 742720/ERC_/European Research Council/International
- 742720/H2020 European Research Council/International
- ID 259139777/Deutsche Forschungsgemeinschaft/International
- DynaMem/LOEWE program of the State of Hesse/International
- (Project-ID259130777 - SFB1177/German Research Foundation/International
LinkOut - more resources
Full Text Sources
Other Literature Sources

