Pedigree-Based Gene Mapping Supports Previous Loci and Reveals Novel Suggestive Loci in Specific Language Impairment
- PMID: 33186502
- PMCID: PMC8608229
- DOI: 10.1044/2020_JSLHR-20-00102
Pedigree-Based Gene Mapping Supports Previous Loci and Reveals Novel Suggestive Loci in Specific Language Impairment
Abstract
Purpose Specific language impairment (SLI) is characterized by a delay in language acquisition despite a lack of other developmental delays or hearing loss. Genetics of SLI is poorly understood. The purpose of this study is to identify SLI genetic loci through family-based linkage mapping. Method We performed genome-wide parametric linkage analysis in six families segregating with SLI. An age-appropriate standardized omnibus language measure was used to categorically define the SLI phenotype. Results A suggestive linkage region replicated a previous region of interest with the highest logarithm of odds (LOD) score of 2.40 at 14q11.2-q13.3 in Family 489. A paternal parent-of-origin effect associated with SLI and language phenotypes on a nonsynonymous single nucleotide polymorphism (SNP) in NOP9 (14q12) was reported previously. Linkage analysis identified a new SLI locus at 15q24.3-25.3 with the highest parametric LOD score of 3.06 in Family 315 under a recessive mode of inheritance. Suggestive evidence of linkage was also revealed at 4q31.23-q35.2 in Family 300, with the highest LOD score of 2.41. Genetic linkage was not identified in the other three families included in parametric linkage analysis. Conclusions These results are the first to report genome-wide suggestive linkage with a total language standard score on an age-appropriate omnibus language measure across a wide age range. Our findings confirm previous reports of a language-associated locus on chromosome 14q, report new SLI loci, and validate the pedigree-based parametric linkage analysis approach to mapping genes for SLI. Supplemental Material https://doi.org/10.23641/asha.13203218.
Figures
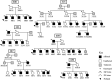

References
-
- Abecasis, G. R. , Cherny, S. S. , Cookson, W. O. , & Cardon, L. R. (2002). Merlin-rapid analysis of dense genetic maps using sparse gene flow trees. Nature Genetics, 30(1), 97–101. https://doi.org/10.1038/ng786 - PubMed
-
- Abreu, P. C. , Greenberg, D. A. , & Hodge, S. E. (1999). Direct power comparisons between simple LOD scores and NPL scores for linkage analysis in complex diseases. The American Journal of Human Genetics, 65(3), 847–857. https://doi.org/10.1086/302536 - PMC - PubMed
-
- Adlof, S. M. , & Hogan, T. P. (2019). If we don't look, we won't see: Measuring language development to inform literacy instruction. Policy Insights from the Behavioral and Brain Sciences, 6(2), 210–217. https://doi.org/10.1177/2372732219839075 - PMC - PubMed
-
- Andres, E. M. , Hafeez, H. , Yousaf, A. , Riazuddin, S. , Rice, M. L. , Basra, M. A. R. , & Raza, M. H. (2019). A genome-wide analysis in consanguineous families reveals new chromosomal loci in specific language impairment (SLI). European Journal of Human Genetics, 27(8), 1274–1285. https://doi.org/10.1038/s41431-019-0398-1 - PMC - PubMed
-
- Bailey-Wilson, J. E. , & Wilson, A. F. (2011). Linkage analysis in the next-generation sequencing era. Human Heredity, 72(4), 228–236. https://doi.org/10.1159/000334381 - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

