Organohalide-Respiring Bacteria at the Heart of Anaerobic Metabolism in Arctic Wet Tundra Soils
- PMID: 33187999
- PMCID: PMC7848920
- DOI: 10.1128/AEM.01643-20
Organohalide-Respiring Bacteria at the Heart of Anaerobic Metabolism in Arctic Wet Tundra Soils
Abstract
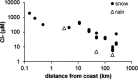
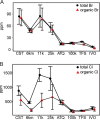
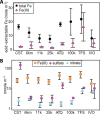
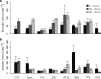
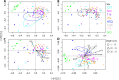
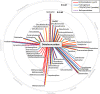
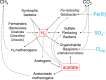
Recent work revealed an active biological chlorine cycle in coastal Arctic tundra of northern Alaska. This raised the question of whether chlorine cycling was restricted to coastal areas or if these processes extended to inland tundra. The anaerobic process of organohalide respiration, carried out by specialized bacteria like Dehalococcoides, consumes hydrogen gas and acetate using halogenated organic compounds as terminal electron acceptors, potentially competing with methanogens that produce the greenhouse gas methane. We measured microbial community composition and soil chemistry along an ∼262-km coastal-inland transect to test for the potential of organohalide respiration across the Arctic Coastal Plain and studied the microbial community associated with Dehalococcoides to explore the ecology of this group and its potential to impact C cycling in the Arctic. Concentrations of brominated organic compounds declined sharply with distance from the coast, but the decrease in organic chlorine pools was more subtle. The relative abundances of Dehalococcoides were similar across the transect, except for being lower at the most inland site. Dehalococcoides correlated with other strictly anaerobic genera, plus some facultative ones, that had the genetic potential to provide essential resources (hydrogen, acetate, corrinoids, or organic chlorine). This community included iron reducers, sulfate reducers, syntrophic bacteria, acetogens, and methanogens, some of which might also compete with Dehalococcoides for hydrogen and acetate. Throughout the Arctic Coastal Plain, Dehalococcoides is associated with the dominant anaerobes that control fluxes of hydrogen, acetate, methane, and carbon dioxide. Depending on seasonal electron acceptor availability, organohalide-respiring bacteria could impact carbon cycling in Arctic wet tundra soils.IMPORTANCE Once considered relevant only in contaminated sites, it is now recognized that biological chlorine cycling is widespread in natural environments. However, linkages between chlorine cycling and other ecosystem processes are not well established. Species in the genus Dehalococcoides are highly specialized, using hydrogen, acetate, vitamin B12-like compounds, and organic chlorine produced by the surrounding community. We studied which neighbors might produce these essential resources for Dehalococcoides species. We found that Dehalococcoides species are ubiquitous across the Arctic Coastal Plain and are closely associated with a network of microbes that produce or consume hydrogen or acetate, including the most abundant anaerobic bacteria and methanogenic archaea. We also found organic chlorine and microbes that can produce these compounds throughout the study area. Therefore, Dehalococcoides could control the balance between carbon dioxide and methane (a more potent greenhouse gas) when suitable organic chlorine compounds are available to drive hydrogen and acetate uptake.
Keywords: Arctic; Dehalococcoides; chlorine; methanogenesis; soil.
Copyright © 2021 American Society for Microbiology.
Figures

References
-
- Zlamal JE, Raab TK, Little M, Edwards RA, Lipson DA. 2017. Biological chlorine cycling in the Arctic Coastal Plain. Biogeochemistry 134:243–260. doi:10.1007/s10533-017-0359-0. - DOI
-
- Löffler FE, Yan J, Ritalahti KM, Adrian L, Edwards EA, Konstantinidis KT, Müller JA, Fullerton H, Zinder SH, Spormann AM. 2013. Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int J Syst Evol Microbiol 63:625–635. doi:10.1099/ijs.0.034926-0. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

