Virus Isoelectric Point Estimation: Theories and Methods
- PMID: 33188001
- PMCID: PMC7848896
- DOI: 10.1128/AEM.02319-20
Virus Isoelectric Point Estimation: Theories and Methods
Abstract
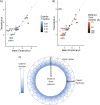
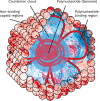
Much of virus fate, both in the environment and in physical/chemical treatment, is dependent on electrostatic interactions. Developing an accurate means of predicting virion isoelectric point (pI) would help to understand and anticipate virus fate and transport, especially for viruses that are not readily propagated in the lab. One simple approach to predicting pI estimates the pH at which the sum of charges from ionizable amino acids in capsid proteins approaches zero. However, predicted pIs based on capsid charges frequently deviate by several pH units from empirically measured pIs. Recently, the discrepancy between empirical and predicted pI was attributed to the electrostatic neutralization of predictable polynucleotide-binding regions (PBRs) of the capsid interior. In this paper, we review models presupposing (i) the influence of the viral polynucleotide on surface charge or (ii) the contribution of only exterior residues to surface charge. We then compare these models to the approach of excluding only PBRs and hypothesize a conceptual electrostatic model that aligns with this approach. The PBR exclusion method outperformed methods based on three-dimensional (3D) structure and accounted for major discrepancies in predicted pIs without adversely affecting pI prediction for a diverse range of viruses. In addition, the PBR exclusion method was determined to be the best available method for predicting virus pI, since (i) PBRs are predicted independently of the impact on pI, (ii) PBR prediction relies on proteome sequences rather than detailed structural models, and (iii) PBR exclusion was successfully demonstrated on a diverse set of viruses. These models apply to nonenveloped viruses only. A similar model for enveloped viruses is complicated by a lack of data on enveloped virus pI, as well as uncertainties regarding the influence of the phospholipid envelope on charge and ion gradients.
Keywords: DNA binding; DNA-binding proteins; RNA binding; RNA-binding proteins; capsid; colloid; electrostatic; modeling; polynucleotide; prediction; predictive model; surface charge; virion; virion structure.
Copyright © 2021 American Society for Microbiology.
Figures

Similar articles
-
Improved Virus Isoelectric Point Estimation by Exclusion of Known and Predicted Genome-Binding Regions.Appl Environ Microbiol. 2020 Nov 10;86(23):e01674-20. doi: 10.1128/AEM.01674-20. Print 2020 Nov 10. Appl Environ Microbiol. 2020. PMID: 32978129 Free PMC article.
-
Isoelectric points of viruses.J Appl Microbiol. 2010 Aug;109(2):388-397. doi: 10.1111/j.1365-2672.2010.04663.x. Epub 2010 Jan 22. J Appl Microbiol. 2010. PMID: 20102425 Review.
-
Controlling the surface charge of simple viruses.PLoS One. 2021 Sep 10;16(9):e0255820. doi: 10.1371/journal.pone.0255820. eCollection 2021. PLoS One. 2021. PMID: 34506491 Free PMC article.
-
Electrostatics and the assembly of an RNA virus.Phys Rev E Stat Nonlin Soft Matter Phys. 2005 Jun;71(6 Pt 1):061928. doi: 10.1103/PhysRevE.71.061928. Epub 2005 Jun 30. Phys Rev E Stat Nonlin Soft Matter Phys. 2005. PMID: 16089786
-
Energies and pressures in viruses: contribution of nonspecific electrostatic interactions.Phys Chem Chem Phys. 2012 Mar 21;14(11):3746-65. doi: 10.1039/c1cp22756d. Epub 2011 Dec 6. Phys Chem Chem Phys. 2012. PMID: 22143065 Review.
Cited by
-
Navigating the Purification Process: Maintaining the Integrity of Replication-Competent Enveloped Viruses.Vaccines (Basel). 2025 Apr 23;13(5):444. doi: 10.3390/vaccines13050444. Vaccines (Basel). 2025. PMID: 40432057 Free PMC article. Review.
-
CrAssphage as a Human Enteric Viral Contamination Bioindicator in Marketed Bivalve Mollusks.Viruses. 2025 Jul 18;17(7):1012. doi: 10.3390/v17071012. Viruses. 2025. PMID: 40733628 Free PMC article.
-
A Roadmap for Building Waterborne Virus Traps.JACS Au. 2022 Oct 4;2(10):2205-2221. doi: 10.1021/jacsau.2c00377. eCollection 2022 Oct 24. JACS Au. 2022. PMID: 36311831 Free PMC article. Review.
-
Mechanisms, Techniques and Devices of Airborne Virus Detection: A Review.Int J Environ Res Public Health. 2023 Apr 11;20(8):5471. doi: 10.3390/ijerph20085471. Int J Environ Res Public Health. 2023. PMID: 37107752 Free PMC article. Review.
-
Development of a Novel Phagomagnetic-Assisted Isothermal DNA Amplification System for Endpoint Electrochemical Detection of Listeria monocytogenes.Biosensors (Basel). 2023 Apr 7;13(4):464. doi: 10.3390/bios13040464. Biosensors (Basel). 2023. PMID: 37185539 Free PMC article.
References
-
- Heffron J, Mayer BK. 2016. Emerging investigators series: virus mitigation by coagulation: recent discoveries and future directions. Environ Sci (Camb) 2:443–459. doi:10.1039/C6EW00060F. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

