FreeClimber: automated quantification of climbing performance in Drosophila
- PMID: 33188065
- PMCID: PMC7823161
- DOI: 10.1242/jeb.229377
FreeClimber: automated quantification of climbing performance in Drosophila
Abstract
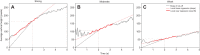
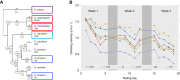
Negative geotaxis (climbing) performance is a useful metric for quantifying Drosophila health. Manual methods to quantify climbing performance are tedious and often biased, while many available computational methods have challenging hardware or software requirements. We present an alternative: FreeClimber. This open source, Python-based platform subtracts a video's static background to improve detection for flies moving across heterogeneous backgrounds. FreeClimber calculates a cohort's velocity as the slope of the most linear portion of a mean vertical position versus time curve. It can run from a graphical user interface for optimization or a command line interface for high-throughput and automated batch processing, improving accessibility for users with different expertise. FreeClimber outputs calculated slopes, spot locations for follow-up analyses (e.g. tracking), and several visualizations and plots. We demonstrate FreeClimber's utility in a longitudinal study for endurance exercise performance in Drosophila mitonuclear genotypes using six distinct mitochondrial haplotypes paired with a common D. melanogaster nuclear background.
Keywords: Automated behavioral analysis; Insect locomotion; Longitudinal performance screen; Python; RING assay; Rapid iterative negative geotaxis assay; Spot detection.
© 2021. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures

References
-
- Bellen H. J., Levis R. W., He Y. C., Carlson J. W., Evans-Holm M., Bae E., Kim J., Metaxakis A., Savakis C., Schulze K. L. et al. (2011). The Drosophila gene disruption project: progress using transposons with distinctive site specificities. Genetics 188, U731-U341 10.1534/genetics.111.126995 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

