Do FeS clusters rule bacterial iron regulation?
- PMID: 33188081
- PMCID: PMC7667982
- DOI: 10.1074/jbc.H120.016190
Do FeS clusters rule bacterial iron regulation?
Abstract
For decades, the bacterial ferric uptake regulator (Fur) has been thought to respond to ferrous iron to transcriptionally regulate genes required for balancing iron uptake, storage, and utilization. Because iron binding to Fur has never been confirmed in vivo, the physiological iron-sensing mechanism remains an open question. Fontenot et al. now show that Fur purified from Escherichia coli binds an all-Cys-coordinated [2Fe-2S] cluster. This finding opens the exciting possibility that Fur may join numerous well-studied bacterial, fungal, and mammalian proteins that use FeS clusters for cellular iron regulation.
© 2020 Lill.
Conflict of interest statement
Conflict of interest—The author declares that he has no conflicts of interest with the contents of this article.
Figures
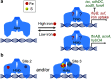
Comment on
-
Ferric uptake regulator (Fur) reversibly binds a [2Fe-2S] cluster to sense intracellular iron homeostasis in Escherichia coli.J Biol Chem. 2020 Nov 13;295(46):15454-15463. doi: 10.1074/jbc.RA120.014814. Epub 2020 Sep 14. J Biol Chem. 2020. PMID: 32928958 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

