Nucleic acid amplification tests on respiratory samples for the diagnosis of coronavirus infections: a systematic review and meta-analysis
- PMID: 33188933
- PMCID: PMC7657614
- DOI: 10.1016/j.cmi.2020.11.002
Nucleic acid amplification tests on respiratory samples for the diagnosis of coronavirus infections: a systematic review and meta-analysis
Abstract
Background: Management and control of coronavirus disease 2019 (COVID-19) relies on reliable diagnostic testing.
Objectives: To evaluate the diagnostic test accuracy (DTA) of nucleic acid amplification tests (NAATs) for the diagnosis of coronavirus infections.
Data sources: PubMed, Web of Science, the Cochrane Library, Embase, Open Grey and conference proceeding until May 2019. PubMed and medRxiv were updated for COVID-19 on 31st August 2020.
Study eligibility: Studies were eligible if they reported on agreement rates between different NAATs using clinical samples.
Participants: Symptomatic patients with suspected upper or lower respiratory tract coronavirus infection.
Methods: The new NAAT was defined as the index test and the existing NAAT as reference standard. Data were extracted independently in duplicate. Risk of bias was assessed using the Quality Assessment of Diagnostic Accuracy Studies 2 tool. Confidence regions (CRs) surrounding summary sensitivity/specificity pooled by bivariate meta-analysis are reported. Heterogeneity was assessed using meta-regression.
Results: Fifty-one studies were included, 22 of which included 10 181 persons before COVID-19 and 29 including 8742 persons diagnosed with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The overall summary sensitivity was 89.1% (95%CR 84.0-92.7%) and specificity 98.9% (95%CR 98.0-99.4%). Nearly all the studies evaluated different PCRs as both index and reference standards. Real-time RT PCR assays resulted in significantly higher sensitivity than other tests. Reference standards at high risk of bias possibly exaggerated specificity. The pooled sensitivity and specificity of studies evaluating SARS-COV-2 were 90.4% (95%CR 83.7-94.5%) and 98.1% (95%CR 95.9-99.2), respectively. SARS-COV-2 studies using samples from the lower respiratory tract, real-time RT-PCR, and tests targeting the N or S gene or more than one gene showed higher sensitivity, and assays based on reverse transcriptase loop-mediated isothermal amplification (RT-LAMP), especially when targeting only the RNA-dependent RNA polymerase (RdRp) gene, showed significantly lower sensitivity compared to other studies.
Conclusions: Pooling all studies to date shows that on average 10% of patients with coronavirus infections might be missed with PCR tests. Variables affecting sensitivity and specificity can be used for test selection and development.
Keywords: Acute respiratory tract infection; COVID-19; Coronavirus; Nucleic acid amplification tests; SARS-CoV-2.
Copyright © 2020 European Society of Clinical Microbiology and Infectious Diseases. Published by Elsevier Ltd. All rights reserved.
Figures
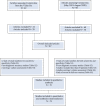
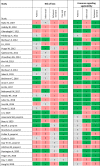



References
-
- Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020 - PubMed
-
- Centers for Disease Control and Prevention . Centers for Disease Control and Prevention; 2020. CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel 2020.https://www.fda.gov/media/134922/download Available from:
-
- Whiting P.F., Rutjes A.W., Westwood M.E., Mallett S., Deeks J.J., Reitsma J.B. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529–536. - PubMed
-
- Reitsma J.B., Glas A.S., Rutjes A.W., Scholten R.J., Bossuyt P.M., Zwinderman A.H. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J Clin Epidemiol. 2005;58:982–990. - PubMed
-
- Schwarzer G., Carpenter J., Rücker G. Springer International Publishing AG; 2015. Meta-analysis with R.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

