Increased H3K4me3 methylation and decreased miR-7113-5p expression lead to enhanced Wnt/β-catenin signaling in immune cells from PTSD patients leading to inflammatory phenotype
- PMID: 33189141
- PMCID: PMC7666486
- DOI: 10.1186/s10020-020-00238-3
Increased H3K4me3 methylation and decreased miR-7113-5p expression lead to enhanced Wnt/β-catenin signaling in immune cells from PTSD patients leading to inflammatory phenotype
Abstract
Background: Posttraumatic stress disorder (PTSD) is a psychiatric disorder accompanied by chronic peripheral inflammation. What triggers inflammation in PTSD is currently unclear. In the present study, we identified potential defects in signaling pathways in peripheral blood mononuclear cells (PBMCs) from individuals with PTSD.
Methods: RNAseq (5 samples each for controls and PTSD), ChIPseq (5 samples each) and miRNA array (6 samples each) were used in combination with bioinformatics tools to identify dysregulated genes in PBMCs. Real time qRT-PCR (24 samples each) and in vitro assays were employed to validate our primary findings and hypothesis.
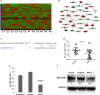
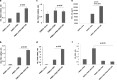
Results: By RNA-seq analysis of PBMCs, we found that Wnt signaling pathway was upregulated in PTSD when compared to normal controls. Specifically, we found increased expression of WNT10B in the PTSD group when compared to controls. Our findings were confirmed using NCBI's GEO database involving a larger sample size. Additionally, in vitro activation studies revealed that activated but not naïve PBMCs from control individuals expressed more IFNγ in the presence of recombinant WNT10B suggesting that Wnt signaling played a crucial role in exacerbating inflammation. Next, we investigated the mechanism of induction of WNT10B and found that increased expression of WNT10B may result from epigenetic modulation involving downregulation of hsa-miR-7113-5p which targeted WNT10B. Furthermore, we also observed that WNT10B overexpression was linked to higher expression of H3K4me3 histone modification around the promotor of WNT10B. Additionally, knockdown of histone demethylase specific to H3K4me3, using siRNA, led to increased expression of WNT10B providing conclusive evidence that H3K4me3 indeed controlled WNT10B expression.
Conclusions: In summary, our data demonstrate for the first time that Wnt signaling pathway is upregulated in PBMCs of PTSD patients resulting from epigenetic changes involving microRNA dysregulation and histone modifications, which in turn may promote the inflammatory phenotype in such cells.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
-
- American Psychiatric Association . DSM-5 Task Force: diagnostic and statistical manual of mental disorders: DSM-5. Washington, D.C.: American Psychiatric Association; 2013.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

