The nucleocapsid protein of zoonotic betacoronaviruses is an attractive target for antiviral drug discovery
- PMID: 33189817
- PMCID: PMC7658559
- DOI: 10.1016/j.lfs.2020.118754
The nucleocapsid protein of zoonotic betacoronaviruses is an attractive target for antiviral drug discovery
Abstract
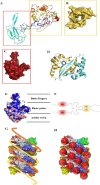
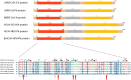
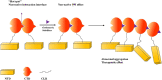
Betacoronaviruses are in one genera of coronaviruses including severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome-related coronavirus (MERS-CoV), etc. These viruses threaten public health and cause dramatic economic losses. The nucleocapsid (N) protein is a structural protein of betacoronaviruses with multiple functions such as forming viral capsids with viral RNA, interacting with viral membrane protein to form the virus core with RNA, binding to several cellular kinases for signal transductions, etc. In this review, we highlighted the potential of the N protein as a suitable antiviral target from different perspectives, including structure, functions, and antiviral strategies for combatting betacoronaviruses.
Keywords: Antiviral; Betacoronavirus; Coronavirus N; Nucleocapsid protein; Protein-protein interaction.
Copyright © 2020. Published by Elsevier Inc.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

Similar articles
-
3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.Sci Transl Med. 2020 Aug 19;12(557):eabc5332. doi: 10.1126/scitranslmed.abc5332. Epub 2020 Aug 3. Sci Transl Med. 2020. PMID: 32747425 Free PMC article.
-
Broad-spectrum coronavirus antiviral drug discovery.Expert Opin Drug Discov. 2019 Apr;14(4):397-412. doi: 10.1080/17460441.2019.1581171. Epub 2019 Mar 8. Expert Opin Drug Discov. 2019. PMID: 30849247 Free PMC article. Review.
-
SARS, MERS and SARS-CoV-2 (COVID-19) treatment: a patent review.Expert Opin Ther Pat. 2020 Aug;30(8):567-579. doi: 10.1080/13543776.2020.1772231. Epub 2020 Jun 7. Expert Opin Ther Pat. 2020. PMID: 32429703 Review.
-
Promising Antiviral Activities of Natural Flavonoids against SARS-CoV-2 Targets: Systematic Review.Int J Mol Sci. 2021 Oct 14;22(20):11069. doi: 10.3390/ijms222011069. Int J Mol Sci. 2021. PMID: 34681727 Free PMC article.
-
Coronavirus Porcine Epidemic Diarrhea Virus Nucleocapsid Protein Interacts with p53 To Induce Cell Cycle Arrest in S-Phase and Promotes Viral Replication.J Virol. 2021 Jul 26;95(16):e0018721. doi: 10.1128/JVI.00187-21. Epub 2021 Jul 26. J Virol. 2021. PMID: 34037422 Free PMC article.
Cited by
-
Characterization and application of a series of monoclonal antibodies against SARS-CoV-2 nucleocapsid protein.J Med Virol. 2023 Jan;95(1):e28225. doi: 10.1002/jmv.28225. Epub 2022 Oct 25. J Med Virol. 2023. PMID: 36238992 Free PMC article.
-
Targeting intracellular Neu1 for coronavirus infection treatment.iScience. 2023 Feb 17;26(2):106037. doi: 10.1016/j.isci.2023.106037. Epub 2023 Jan 24. iScience. 2023. PMID: 36714013 Free PMC article.
-
The Role of NEU1 in Coronavirus Infection and Pathogenesis.Virol Immunol J. 2024;8(3):351. doi: 10.23880/vij-16000351. Epub 2024 Aug 8. Virol Immunol J. 2024. PMID: 40786010 Free PMC article.
-
Multivalent binding of the partially disordered SARS-CoV-2 nucleocapsid phosphoprotein dimer to RNA.Biophys J. 2021 Jul 20;120(14):2890-2901. doi: 10.1016/j.bpj.2021.03.023. Epub 2021 Mar 29. Biophys J. 2021. PMID: 33794152 Free PMC article.
-
Mass Spectrometry Analysis of SARS-CoV-2 Nucleocapsid Protein Reveals Camouflaging Glycans and Unique Post-Translational Modifications.Infect Microbes Dis. 2021 Aug 3;3(3):149-157. doi: 10.1097/IM9.0000000000000071. eCollection 2021 Sep. Infect Microbes Dis. 2021. PMID: 38630108 Free PMC article.
References
-
- The International Committee for Taxonomy of Viruses (ICTV) [(accessed on 27 June 2014)]. Available online: http://talk.ictvonline.org/files/ictv_documents/m/msl/4090.aspx.
-
- Enjuanes Luis., editor. Coronavirus Replication and Reverse Genetics. vol. 287. Springer Science & Business Media; 2004.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

