Integration of whole-exome and anchored PCR-based next generation sequencing significantly increases detection of actionable alterations in precision oncology
- PMID: 33190043
- PMCID: PMC7674614
- DOI: 10.1016/j.tranon.2020.100944
Integration of whole-exome and anchored PCR-based next generation sequencing significantly increases detection of actionable alterations in precision oncology
Abstract
Background: Frequency of clinically relevant mutations in solid tumors by targeted and whole-exome sequencing is ∼30%. Transcriptome analysis complements detection of actionable gene fusions in advanced cancer patients. Goal of this study was to determine the added value of anchored multiplex PCR (AMP)-based next-generation sequencing (NGS) assay to identify further potential drug targets, when coupled with whole-exome sequencing (WES).
Methods: Selected series of fifty-six samples from 55 patients enrolled in our precision medicine study were interrogated by WES and AMP-based NGS. RNA-seq was performed in 19 cases. Clinically relevant and actionable alterations detected by three methods were integrated and analyzed.
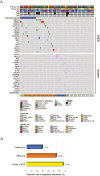
Results: AMP-based NGS detected 48 fusions in 31 samples (55.4%); 31.25% (15/48) were classified as targetable based on published literature. WES revealed 29 samples (51.8%) harbored targetable alterations. TMB-high and MSI-high status were observed in 12.7% and 1.8% of cases. RNA-seq from 19 samples identified 8 targetable fusions (42.1%), also captured by AMP-based NGS. When number of actionable fusions detected by AMP-based NGS were added to WES targetable alterations, 66.1% of samples had potential drug targets. When both WES and RNA-seq were analyzed, 57.8% of samples had targetable alterations.
Conclusions: This study highlights importance of an integrative genomic approach for precision oncology, including use of different NGS platforms with complementary features. Integrating RNA data (whole transcriptome or AMP-based NGS) significantly enhances detection of potential targets in cancer patients. In absence of fresh frozen tissue, AMP-based NGS is a robust method to detect actionable fusions using low-input RNA from archival tissue.
Keywords: Anchored multiplex PCR-based next-generation sequencing; Novel fusion; Oncogenic; RNA Sequencing; Whole-exome sequencing.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare no conflict of interest.
Figures


References
-
- Andre F. Comparative genomic hybridisation array and DNA sequencing to direct treatment of metastatic breast cancer: a multicentre, prospective trial (SAFIR01/UNICANCER) Lancet Oncol. 2014;15(3):267–274. - PubMed
-
- Massard C. High-throughput genomics and clinical outcome in hard-to-treat advanced cancers: results of the MOSCATO 01 trial. Cancer Discov. 2017;7(6):586–595. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

