High-throughput fetal fraction amplification increases analytical performance of noninvasive prenatal screening
- PMID: 33190143
- PMCID: PMC7935715
- DOI: 10.1038/s41436-020-01009-5
High-throughput fetal fraction amplification increases analytical performance of noninvasive prenatal screening
Abstract
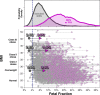
Purpose: The percentage of a maternal cell-free DNA (cfDNA) sample that is fetal-derived (the fetal fraction; FF) is a key driver of the sensitivity and specificity of noninvasive prenatal screening (NIPS). On certain NIPS platforms, >20% of women with high body mass index (and >5% overall) receive a test failure due to low FF (<4%).
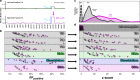
Methods: A scalable fetal fraction amplification (FFA) technology was analytically validated on 1264 samples undergoing whole-genome sequencing (WGS)-based NIPS. All samples were tested with and without FFA.
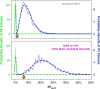
Results: Zero samples had FF < 4% when screened with FFA, whereas 1 in 25 of these same patients had FF < 4% without FFA. The average increase in FF was 3.9-fold for samples with low FF (2.3-fold overall) and 99.8% had higher FF with FFA. For all abnormalities screened on NIPS, z-scores increased 2.2-fold on average in positive samples and remained unchanged in negative samples, powering an increase in NIPS sensitivity and specificity.
Conclusion: FFA transforms low-FF samples into high-FF samples. By combining FFA with WGS-based NIPS, a single round of NIPS can provide nearly all women with confident results about the broad range of potential fetal chromosomal abnormalities across the genome.
Keywords: analytical validation; body mass index; cell-free DNA; fetal fraction; noninvasive prenatal screening.
Figures

References
-
- Palomaki GE, Kloza EM, Lambert-Messerlian GM, et al. DNA sequencing of maternal plasma to detect Down syndrome: an international clinical validation study. Genet Med. 2011;13:913–920. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

