Protease targeted COVID-19 drug discovery and its challenges: Insight into viral main protease (Mpro) and papain-like protease (PLpro) inhibitors
- PMID: 33191083
- PMCID: PMC7647411
- DOI: 10.1016/j.bmc.2020.115860
Protease targeted COVID-19 drug discovery and its challenges: Insight into viral main protease (Mpro) and papain-like protease (PLpro) inhibitors
Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) brutally perils physical and mental health worldwide. Unavailability of effective anti-viral drug rendering global threat of COVID-19 caused by SARS-CoV-2. In this scenario, viral protease enzymes are crucial targets for drug discovery. This extensive study meticulously focused on two viral proteases such as main protease (Mpro) and papain-like protease (PLpro), those are essential for viral replication. This review provides a detail overview of the targets (Mpro and PLpro) from a structural and medicinal chemistry point of view, together with recently reported protease inhibitors. An insight into the challenges in the development of effective as well as drug like protease inhibitors is discussed. Peptidomimetic and/or covalent coronavirus protease inhibitors possessed potent and selective active site inhibition but compromised in pharmacokinetic parameters to be a drug/drug like molecule. Lead optimization of non-peptidomimetic and/or low molecular weight compounds may be a better option for oral delivery. A masterly combination of adequate pharmacokinetic properties with coronavirus protease activity as well as selectivity will provide potential drug candidates in future. This study is a part of our endeavors which surely dictates medicinal chemistry efforts to discover effective anti-viral agent for this devastating disease.
Keywords: COVID-19; Mpro; Non-covalent inhibitor; PLpro; SARS-CoV-2; Structure-activity relationship (SAR).
Copyright © 2020 Elsevier Ltd. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



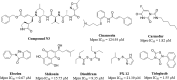

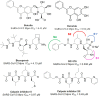

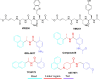
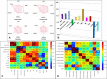
References
-
- Jin Z., Du X., Xu Y. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature. 2020;582:289–293. - PubMed
-
- Zhang L., Lin D., Kusov Y. α-Ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication: structure-based design, synthesis, and activity assessment. J Med Chem. 2020;63:4562–4578. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Miscellaneous

