Microbial population dynamics during traditional production of Mabisi, a spontaneous fermented milk product from Zambia: a field trial
- PMID: 33191438
- PMCID: PMC7667141
- DOI: 10.1007/s11274-020-02957-5
Microbial population dynamics during traditional production of Mabisi, a spontaneous fermented milk product from Zambia: a field trial
Abstract
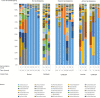
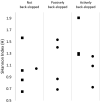
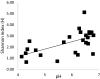
Mabisi is a fermented milk product, traditionally produced in a calabash by uncontrolled fermentation. Due to high costs and a reduced availability of calabashes, nowadays plastic containers are also used for Mabisi production. However, the effect of this change in production practice on the properties of the product has not been documented. Therefore, we aimed at determining the effect of fermentation vessels and types of back-slopping on acidification and microbial communities during fermentation. A series of fifteen experiments using two types of fermentation vessels (plastic buckets and calabashes) in combination with different types of back-slopping (no back-slopping, passive back-slopping, and active back-slopping) were set up at a field site in rural Zambia. In each of the fifteen fermentations we analysed acidification rate of traditional Mabisi fermentation and bacterial diversity over time. No significant difference was found in terms of microbial diversity during and at the end of fermentation between fermentations performed in buckets or previously used calabashes. Bacterial communities in general decreased in diversity over time, where the drop in pH correlated with a decrease in Shannon Index. In case of active back-slopping, the pH drop started right after inoculation while in the no back-slopping and passive back-slopping fermentations, there was a clear lag phase before acidification started. All experimental series resulted in a microbial community dominated by Lactococcus lactis and a Shannon Index, as a measure for diversity, between 0.6 and 2.0. The use of plastic buckets for Mabisi fermentation can be a valuable alternative to the use of calabashes as this study showed no biological and physico-chemical differences between Mabisi resulting from both fermentation vessels, although the reason for perceived differences should be further investigated.
Keywords: Bacterial community; Dairy; Fermentation vessel; Field study; Traditional fermentation.
Conflict of interest statement
All research is performed with respect to international regulations and scientific ethical practices. The authors declare no conflict of interest. Ethical clearance for this study was obtained from ethics committees at the University of Zambia and at Wageningen University.
Figures






References
-
- Chileshe J, Talsma EF, Schoustra SE, Borgonjen-van den Berg KJ, Handema R, Zwaan BJ, Brouwer ID. Potential contribution of cereal and milk based fermented foods to dietary nutrient intake of 1–5 years old children in Central province in Zambia. PLoS ONE. 2020;15:e0232824. doi: 10.1371/journal.pone.0232824. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

