Comprehensive Analysis of Aberrantly Expressed Competitive Endogenous RNA Network and Identification of Prognostic Biomarkers in Pheochromocytoma and Paraganglioma
- PMID: 33192072
- PMCID: PMC7654541
- DOI: 10.2147/OTT.S271417
Comprehensive Analysis of Aberrantly Expressed Competitive Endogenous RNA Network and Identification of Prognostic Biomarkers in Pheochromocytoma and Paraganglioma
Abstract
Background: Long non-coding RNA (lncRNA) functions as a competitive endogenous RNA (ceRNA) and plays an important role in the biological processes underlying tumorigenesis. However, studies describing the function of lncRNA in pheochromocytoma and paraganglioma (PCPG) remain largely unknown. Our study aims to construct a regulatory ceRNA network and explore prognostic biomarkers for PCPG through a comprehensive analysis.
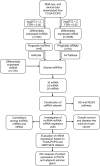
Methods: PCPG data from The Cancer Genome Atlas (TCGA) were utilized to obtain differentially expressed lncRNAs (DElncRNAs), microRNAs (DEmiRNAs), and mRNAs (DEmRNAs). Kaplan-Meier analysis was used to detect prognostic biomarkers and Cytoscape was utilized to construct a regulatory network of ceRNA. Potential lncRNA-miRNA-mRNA axes were inferred by correlation analysis. GO and KEGG pathways were constructed using "clusterProfiler" and "DOSE" R-packages. Immunohistochemistry (IHC) staining was performed to validate differential protein expression levels of genes in the axes. Finally, the GSE19422 dataset and Pan-Cancer data were applied to validate the expression pattern and survival status of mRNAs, respectively.
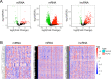
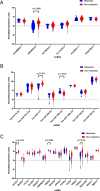
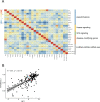
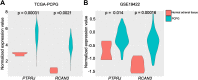
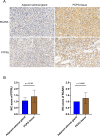
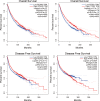
Results: A total of 334 DElncRNAs, 116 DEmiRNAs, and 3496 DEmRNAs were identified and mainly enriched in hormone secretion, metabolism signaling, metastatic and proliferative pathways. Among these differentially expressed genes, 16 mRNAs, six lncRNAs, and two miRNAs were associated with overall survival of patients with PCPG and sequentially enrolled in the ceRNA network. Two lncRNA-miRNA-mRNA regulatory axes were predicted: AP001486.2/hsa-miR-195-5p/RCAN3 and AP006333.2/hsa-miR-34a-5p/PTPRJ. The GSE19422 dataset and IHC analysis validated that mRNA and protein levels of RCAN3 and PTPRJ were upregulated in PCPG tissues compared with adjacent adrenal gland medulla tissues. Pan-Cancer data showed that the upregulated expression of RCAN3 and PTPRJ was associated with favorable overall survival and disease-free survival.
Conclusion: A regulatory lncRNA-miRNA-mRNA ceRNA network was successfully constructed and 24 prognostic biomarkers were identified for PCPG patients. These findings may contribute toward a better understanding of the biological mechanism of tumorigenesis and enable further evaluation of the prognosis of patients with PCPG.
Keywords: ceRNA network; pheochromocytoma and paraganglioma (PCPG); prognostic biomarker.
© 2020 Wang et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures






References
LinkOut - more resources
Full Text Sources

