Genome Editing for CNS Disorders
- PMID: 33192264
- PMCID: PMC7642486
- DOI: 10.3389/fnins.2020.579062
Genome Editing for CNS Disorders
Erratum in
-
Corrigendum: Genome Editing for CNS Disorders.Front Neurosci. 2021 May 26;15:698879. doi: 10.3389/fnins.2021.698879. eCollection 2021. Front Neurosci. 2021. PMID: 34122005 Free PMC article.
Abstract
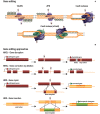
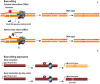
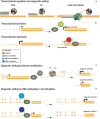
Central nervous system (CNS) disorders have a social and economic burden on modern societies, and the development of effective therapies is urgently required. Gene editing may prevent or cure a disease by inducing genetic changes at endogenous loci. Genome editing includes not only the insertion, deletion or replacement of nucleotides, but also the modulation of gene expression and epigenetic editing. Emerging technologies based on ZFs, TALEs, and CRISPR/Cas systems have extended the boundaries of genome manipulation and promoted genome editing approaches to the level of promising strategies for counteracting genetic diseases. The parallel development of efficient delivery systems has also increased our access to the CNS. In this review, we describe the various tools available for genome editing and summarize in vivo preclinical studies of CNS genome editing, whilst considering current limitations and alternative approaches to overcome some bottlenecks.
Keywords: CNS; CRISPR/Cas; TALEs; ZFs; genome editing.
Copyright © 2020 Duarte and Déglon.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources

