Sphingosine 1-Phosphate Induces Cyclooxygenase-2/Prostaglandin E2 Expression via PKCα-dependent Mitogen-Activated Protein Kinases and NF-κB Cascade in Human Cardiac Fibroblasts
- PMID: 33192511
- PMCID: PMC7662885
- DOI: 10.3389/fphar.2020.569802
Sphingosine 1-Phosphate Induces Cyclooxygenase-2/Prostaglandin E2 Expression via PKCα-dependent Mitogen-Activated Protein Kinases and NF-κB Cascade in Human Cardiac Fibroblasts
Abstract
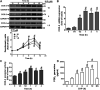
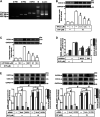
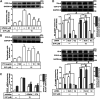
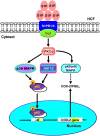
In the regions of tissue injuries and inflammatory diseases, sphingosine 1-phosphate (S1P), a proinflammatory mediator, is increased. S1P may induce the upregulation of cyclooxygenase-2 (COX-2)/prostaglandin E2 (PGE2) system in various types of cells to exacerbate heart inflammation. However, the detailed molecular mechanisms by which S1P induces COX-2 expression in human cardiac fibroblasts (HCFs) remain unknown. HCFs were incubated with S1P and analyzed by Western blotting, real time-Polymerase chain reaction (RT-PCR), and immunofluorescent staining. Our results indicated that S1P activated S1PR1/3-dependent transcriptional activity to induce COX-2 expression and PGE2 production. S1P recruited and activated PTX-sensitive Gi or -insensitive Gq protein-coupled S1PR and then stimulated PKCα-dependent phosphorylation of p42/p44 MAPK, p38 MAPK, and JNK1/2, leading to activating transcription factor NF-κB. Moreover, S1P-activated NF-κB was translocated into the nucleus and bound to its corresponding binding sites on COX-2 promoters determined by chromatin immunoprecipitation (ChIP) and promoter-reporter assays, thereby turning on COX-2 gene transcription associated with PGE2 production in HCFs. These results concluded that in HCFs, activation of NF-κB by PKCα-mediated MAPK cascades was essential for S1P-induced up-regulation of the COX-2/PGE2 system. Understanding the mechanisms of COX-2 expression and PGE2 production regulated by the S1P/S1PRs system on cardiac fibroblasts may provide rationally therapeutic interventions for heart injury or inflammatory diseases.
Keywords: NF-κB; PKCα; cyclooxygenase-2; human cardiac fibroblasts; mitogen-activated protein kinases; sphingosine 1-phosphate.
Copyright © 2020 Yang, Hsiao, Su and Yang.
Figures






References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

