Quorum-Sensing Regulator OpaR Directly Represses Seven Protease Genes in Vibrio parahaemolyticus
- PMID: 33193123
- PMCID: PMC7658014
- DOI: 10.3389/fmicb.2020.534692
Quorum-Sensing Regulator OpaR Directly Represses Seven Protease Genes in Vibrio parahaemolyticus
Abstract
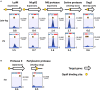
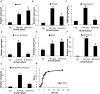
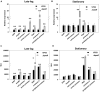
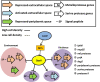
Proteases play a key role in numerous bacterial physiological events. Microbial proteases are used in the pharmaceutical industry and in biomedical applications. The genus Vibrio comprises protease-producing bacteria. Proteases transform polypeptides into shorter chains for easier utilization. They also function as a virulence factor in pathogens. The mechanism by which protease genes are regulated in Vibrio parahaemolyticus, an emerging world-wide human pathogen, however, still remains unclear. Quorum sensing is the communication system of bacteria. OpaR is the master quorum-sensing regulator in V. parahaemolyticus. In the present study, quantitative reverse transcriptase-polymerase chain reaction and protease gene promoter-fusion reporter assays revealed that OpaR represses seven protease genes-three metalloprotease genes and four serine protease genes-which are involved in environmental survival and bacterial virulence. Furthermore, the electrophoresis mobility shift assay demonstrated that OpaR is bound directly to the promoter region of each of the seven protease genes. DNase I footprinting identified the sequence of these OpaR-binding sites. ChIP-seq analyses revealed 435 and 835 OpaR-binding sites in the late-log and stationary phases, respectively. These OpaR-binding sequences indicated a conserved OpaR-binding motif: TATTGATAAAATTATCAATA. These results advance our understanding of the protease regulation system in V. parahaemolyticus. This study is the first to reveal the OpaR motif within V. parahaemolyticus in vivo, using ChIP-seq, and to provide a database for OpaR direct regulon.
Keywords: ChIP-seq; DNase I footprinting; EMSA; OpaR-binding motif; Vibrio parahaemolyticus; proteases regulation; quorum sensing.
Copyright © 2020 Chang and Lee.
Figures



References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

