The Clustered Regularly Interspaced Short Palindromic Repeat System and Argonaute: An Emerging Bacterial Immunity System for Defense Against Natural Transformation?
- PMID: 33193265
- PMCID: PMC7642515
- DOI: 10.3389/fmicb.2020.593301
The Clustered Regularly Interspaced Short Palindromic Repeat System and Argonaute: An Emerging Bacterial Immunity System for Defense Against Natural Transformation?
Abstract
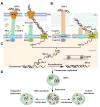
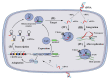
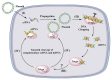
Clustered regularly interspaced short palindromic repeat (CRISPR) systems and prokaryotic Argonaute proteins (Agos) have been shown to defend bacterial and archaeal cells against invading nucleic acids. Indeed, they are important elements for inhibiting horizontal gene transfer between bacterial and archaeal cells. The CRISPR system employs an RNA-guide complex to target invading DNA or RNA, while Agos target DNA using single stranded DNA or RNA as guides. Thus, the CRISPR and Agos systems defend against exogenous nucleic acids by different mechanisms. It is not fully understood how antagonization of these systems occurs during natural transformation, wherein exogenous DNA enters a host cell as single stranded DNA and is then integrated into the host genome. In this review, we discuss the functions and mechanisms of the CRISPR system and Agos in cellular defense against natural transformation.
Keywords: Argonaute proteins; Clustered regularly interspaced short palindromic repeat-Cas; bacterial immunity system; natural transformation; ssDNA.
Copyright © 2020 Liu, Huang, Wang, Zhu, Jia, Chen, Zhang, Pan and Cheng.
Figures
Similar articles
-
Role of CRISPR/cas system in the development of bacteriophage resistance.Adv Virus Res. 2012;82:289-338. doi: 10.1016/B978-0-12-394621-8.00011-X. Adv Virus Res. 2012. PMID: 22420856 Review.
-
Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR) RNAs in the Porphyromonas gingivalis CRISPR-Cas I-C System.J Bacteriol. 2017 Oct 31;199(23):e00275-17. doi: 10.1128/JB.00275-17. Print 2017 Dec 1. J Bacteriol. 2017. PMID: 28893837 Free PMC article.
-
A bacterial Argonaute with noncanonical guide RNA specificity.Proc Natl Acad Sci U S A. 2016 Apr 12;113(15):4057-62. doi: 10.1073/pnas.1524385113. Epub 2016 Mar 30. Proc Natl Acad Sci U S A. 2016. PMID: 27035975 Free PMC article.
-
Postreplication targeting of transformants by bacterial immune systems?Trends Microbiol. 2013 Oct;21(10):516-21. doi: 10.1016/j.tim.2013.08.002. Epub 2013 Sep 8. Trends Microbiol. 2013. PMID: 24021553
-
The CRISPR-Cas Immune System and Genetic Transfers: Reaching an Equilibrium.Microbiol Spectr. 2015 Feb;3(1):PLAS-0034-2014. doi: 10.1128/microbiolspec.PLAS-0034-2014. Microbiol Spectr. 2015. PMID: 26104549 Review.
Cited by
-
Argonaute CSR-1A promotes H3K9me3 maintenance to protect somatic development in offspring.Nucleic Acids Res. 2025 Feb 27;53(5):gkaf127. doi: 10.1093/nar/gkaf127. Nucleic Acids Res. 2025. PMID: 40036504 Free PMC article.
-
The Transformation Experiment of Frederick Griffith II: Inclusion of Cellular Heredity for the Creation of Novel Microorganisms.Bioengineering (Basel). 2025 May 15;12(5):532. doi: 10.3390/bioengineering12050532. Bioengineering (Basel). 2025. PMID: 40428151 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources

