Naturally Occurring tRNAs With Non-canonical Structures
- PMID: 33193279
- PMCID: PMC7609411
- DOI: 10.3389/fmicb.2020.596914
Naturally Occurring tRNAs With Non-canonical Structures
Abstract
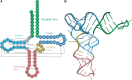
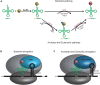
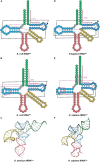
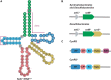
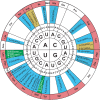
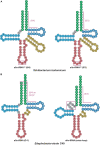
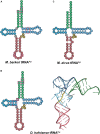
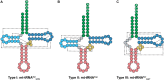
Transfer RNA (tRNA) is the central molecule in genetically encoded protein synthesis. Most tRNA species were found to be very similar in structure: the well-known cloverleaf secondary structure and L-shaped tertiary structure. Furthermore, the length of the acceptor arm, T-arm, and anticodon arm were found to be closely conserved. Later research discovered naturally occurring, active tRNAs that did not fit the established 'canonical' tRNA structure. This review discusses the non-canonical structures of some well-characterized natural tRNA species and describes how these structures relate to their role in translation. Additionally, we highlight some newly discovered tRNAs in which the structure-function relationship is not yet fully understood.
Keywords: genetic code expansion; identity elements; mitochondria; non-canonical; pyrrolysine; selenocysteine; tRNA; translation.
Copyright © 2020 Krahn, Fischer and Söll.
Figures


References
-
- Baron C., Böck A. (1991). The length of the aminoacyl-acceptor stem of the selenocysteine-specific tRNASec of Escherichia coli is the determinant for binding to elongation factors SELB or Tu. J. Biol. Chem. 266 20375–20379. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

