Changing Expression Profiles of Messenger RNA, MicroRNA, Long Non-coding RNA, and Circular RNA Reveal the Key Regulators and Interaction Networks of Competing Endogenous RNA in Pulmonary Fibrosis
- PMID: 33193637
- PMCID: PMC7541945
- DOI: 10.3389/fgene.2020.558095
Changing Expression Profiles of Messenger RNA, MicroRNA, Long Non-coding RNA, and Circular RNA Reveal the Key Regulators and Interaction Networks of Competing Endogenous RNA in Pulmonary Fibrosis
Abstract
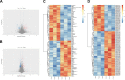
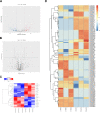
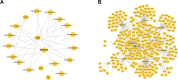
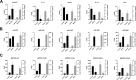
Pulmonary fibrosis is a kind of interstitial lung disease with architectural remodeling of tissues and excessive matrix deposition. Apart from messenger RNA (mRNA), microRNA (miRNA), long non-coding RNA (lncRNA), and circular RNA (circRNA) could also play important roles in the regulatory processes of occurrence and progression of pulmonary fibrosis. In the present study, the pulmonary fibrosis model was administered with bleomycin. Whole transcriptome sequencing analysis was applied to investigate the expression profiles of mRNAs, lncRNAs, circRNAs, and miRNAs. After comparing bleomycin-induced pulmonary fibrosis model lung samples and controls, 286 lncRNAs, 192 mRNAs, 605 circRNAs, and 32 miRNAs were found to be differentially expressed. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed to investigate the potential functions of these differentially expressed (DE) mRNAs and non-coding RNAs (ncRNAs). The terms related to inflammatory response and tumor necrosis factor (TNF) signaling pathway were enriched, implying potential roles in regulatory process. In addition, two co-expression networks were also constructed to understand the internal regulating relationships of these mRNAs and ncRNAs. Our study provides a systematic perspective on the potential functions of these DE mRNAs and ncRNAs during PF process and could help pave the way for effective therapeutics for this devastating and complex disease.
Keywords: ceRNA; co-expression network; pulmonary fibrosis; regulation; whole transcriptome sequencing.
Copyright © 2020 Liu, Liu, Jia, He, Zhang and Zhang.
Figures


References
-
- Ambros V. (2001). microRNAs: tiny regulators with great potential. Cell 107 823–826. - PubMed
LinkOut - more resources
Full Text Sources

