Genome-Wide Assessment of DNA Methylation in Chicken Cardiac Tissue Exposed to Different Incubation Temperatures and CO2 Levels
- PMID: 33193638
- PMCID: PMC7655987
- DOI: 10.3389/fgene.2020.558189
Genome-Wide Assessment of DNA Methylation in Chicken Cardiac Tissue Exposed to Different Incubation Temperatures and CO2 Levels
Abstract
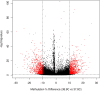
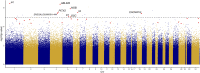
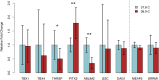
Temperature and CO2 concentration during incubation have profound effects on broiler chick development, and numerous studies have identified significant effects on hatch heart weight (HW) as a result of differences in these parameters. Early life environment has also been shown to affect broiler performance later in life; it has thus been suggested that epigenetic mechanisms may mediate long-term physiological changes induced by environmental stimuli. DNA methylation is an epigenetic modification that can confer heritable changes in gene expression. Using reduced-representation bisulfite sequencing (RRBS), we assessed DNA methylation patterns in cardiac tissue of 84 broiler hatchlings incubated at two egg shell temperatures (EST; 37.8°C and 38.9°C) and three CO2 concentrations (0.1%, 0.4%, and 0.8%) from day 8 of incubation onward. We assessed differential methylation between EST treatments and identified 2,175 differentially methylated (DM) CpGs (1,121 hypermethylated, 1,054 hypomethylated at 38.9° vs. 37.8°) in 269 gene promoters and 949 intragenic regions. DM genes (DMGs) were associated with heart developmental processes, including cardiomyocyte proliferation and differentiation. We identified enriched binding motifs among DM loci, including those for transcription factors associated with cell proliferation and heart development among hypomethylated CpGs that suggest increased binding ability at higher EST. We identified 9,823 DM CpGs between at least two CO2 treatments, with the greatest difference observed between 0.8 and 0.1% CO2 that disproportionately impacted genes involved in cardiac muscle development and response to low oxygen levels. Using HW measurements from the same chicks, we performed an epigenome-wide association study (EWAS) for HW, and identified 23 significantly associated CpGs, nine of which were also DM between ESTs. We found corresponding differences in transcript abundance between ESTs in three DMGs (ABLIM2, PITX2, and THRSP). Hypomethylation of an exonic CpG in PITX2 at 38.9°C was associated with increased expression, and suggests increased cell proliferation in broiler hatchlings incubated at higher temperatures. Overall, these results identified numerous epigenetic associations between chick incubation factors and heart development that may manifest in long-term differences in animal performance.
Keywords: CO2; DNA methylation; chicken; epigenetics; heart; temperature.
Copyright © 2020 Corbett, te Pas, van den Brand, Groenen, Crooijmans, Ernst and Madsen.
Figures

References
LinkOut - more resources
Full Text Sources
Research Materials

